Figures & data
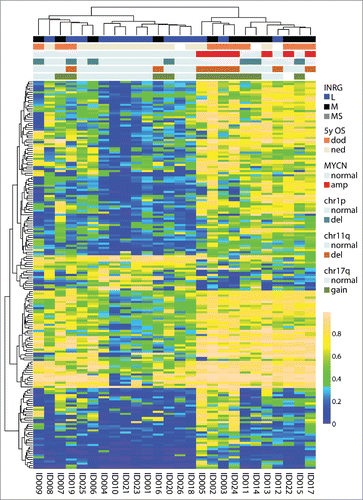
Figure 1. Top 1% most variable sites. (A) Clustering dendrogram from hierarchical clustering. Differences between clusters were tested with χ2 statistics (except for chr11q and chr12 that were tested with Fisher exact test due to low numbers). Abbreviations: amp, amplified; del, deletion; OS_5y, Overall survival 5 years; dod, dead of disease; ned, no evidence of disease (>60 months; samples with ned <60 months were blanked in display); chr1p, chromosome 1p; chr11q, chromosome 11q; chr12, chromosome 12; chr17q, chromosome 17q; ns, non-significant. (B) Boxplot presenting mean methylation level in cluster 1 compared with cluster 2 (P-value < 2.2×10−16, Student t-test). Upper and lower hinges of the box represent the 75th percentile and 25th percentile respectively; whiskers indicate the highest and lowest values that are not outliers; thick horizontal line within box, median. Open circles represent outliers. (C) Kaplan-Meier plot representing survival probability of cluster 1 and cluster 2. (P=0.011; log-rank test).
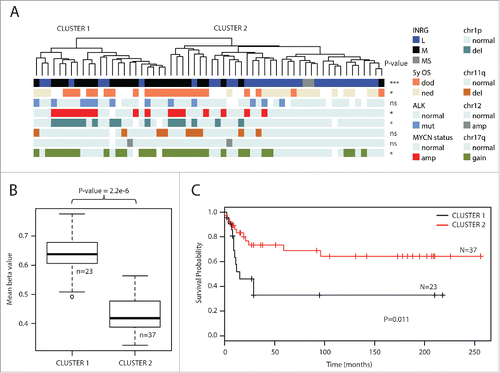
Figure 2. INRG methylated variable positions. (A) Volcano plot showing the distribution between adjusted P-values and difference in β value (deltaBeta) from the ChAMP analysis of the 450K data of INRG M (n=26) and L (n=32) samples. Lines represent used cut-off values to identify the most hyper- and hypomethylated sites in INRG M (n=4557). (B) Distribution of hyper- and hypomethylated loci (MVP; methylated variable position) across CpG features (top) and gene regions (bottom). (C) The 3 INRG M hyper and hypo MVPs with smallest P-values. (D) Kaplan–Meier plots showing survival probability for subjects with above median methylation (red line; n=30), and below median methylation (black line; n=30) for the 3 INRG hyper, and hypo MVPs with lowest P-values. P-value of log-rank test is shown in each plot.
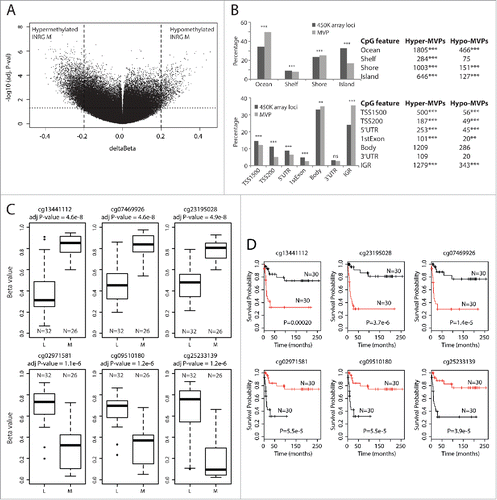
Figure 3. Hypermethylated genes in INRG stage (M) compared to stage L. (A) Gene plot of the TERT gene. The TERT gene represented by a UCSC track (http://genome.ucsc.edu/),Citation32 showing TERT transcripts and positions of 450K probes (upper panel). Gene plot showing mean methylation levels of INRG M, and L for all probes on the Illumina 450K platform annotated to TERT gene (lower panel); (B) The 10 gene ontologies (GO) with lowest P-value from functional GO analysis using the DAVID software. Adj P-value; Benjamini adjusted P-value. (C) Chromosomal distribution of genes at INRG DMRs.
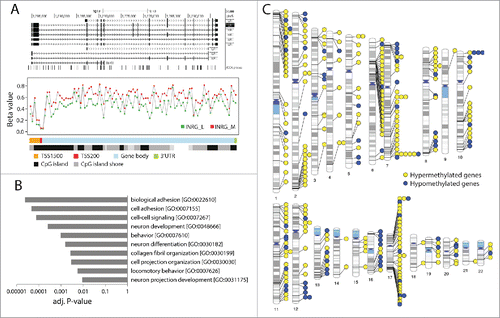
Table 1. Genes with highest number of hyper or hypo MVPs in INRG M group.
Table 2. The strongest DMRs when comparing INRG groups.
Figure 4. Non-CpG methylation. (A) Hierarchical cluster analysis of most variable non-CpG sites (n=168 sites) defines 3 main groups of patients with different methylation frequency. (B) Boxplot presenting mean methylation level of non-CpG sites in INRG L (n=32) compared with INRG M (n=26).
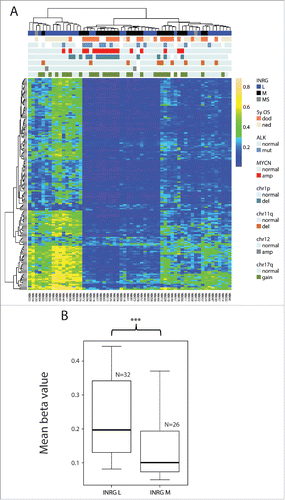
Figure 5. Validation of the INRG separation from the 450K results with 27K data. 187 probes from the intersect that were on both the 450K and the 27K platform separated the samples on cluster 1 and cluster 2 (both on INRG and survival, 2-sided Fisher exact probability test P=0.045 and P=0.013, respectively). OS, overall survival; dod, dead of disease; ned, no evidence of disease; amp, amplified; del, deletion.