Figures & data
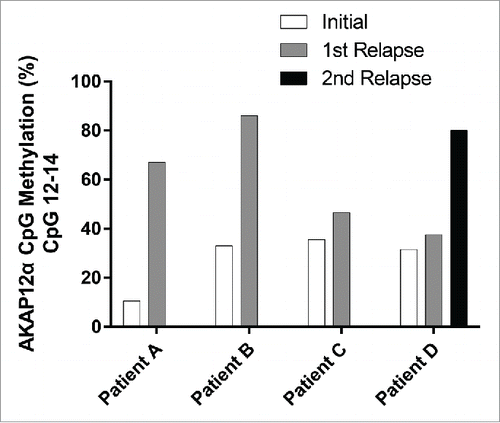
Figure 1. Quantitative DNA methylation analysis of the AKAP12α, β, and γ promoters in JMML. (A) Schematic representation of the AKAP12 locus showing the promoter regions of AKAP12α-, β- and γ-isoforms with their respective CpG islands. All isoforms share a common N-terminus encoded by 2 exons (C2 and C3). CpG islands are depicted as gray boxes (<300 bp, light gray). The MassARRAY amplicons (gray lines) and their positions relative to their respective TSS are illustrated underneath the CpG islands: α1: −868to −438 bp from TSS, α2: −462 to −131 bp from TSS, β1: −193 to +267 bp from TSS, γ1: −243 to +229 bp from TSS. Methylation heat maps are shown for each amplicon. Each row represents a sample and each column a CpG unit. Percentage of DNA methylation ranges from 0% (light green) to 100% (dark blue). Gray indicates unavailable data. HG: healthy granulocytes. (B) The mean DNA methylation levels for each amplicon are displayed as dot plots. The methylation levels between JMML samples (circles) and healthy granulocytes (diamonds) were compared using an unpaired t test with Welch's correction. (C) Leukemic cells isolated from bone marrow (BM) and peripheral blood (PB) were obtained from three JMML patients. DNA methylation values measured for the AKAP12 locus (amplicons α1, α2, β and γ) show a very good correlation between the two tissues (r = 0.996). (D) The methylation profile of amplicon α1 at CpG-unit resolution is shown for JMML and HG. JMML cases were grouped according to their methylation into 25% quartiles. The lower quartile (LQ) contains hypomethylated JMML samples (dashed black line) and the upper quartile (UQ) represents the hypermethylated ones (black line). The average methylation profile of HG (n = 23) is depicted as a light gray line. (E) Methylation levels for CpG units 12, 13, and 14 in AKAP12 amplicon α1 are depicted for HG and all JMML samples, as well as for hypomethylated (LQ) and hypermethylated (HQ) JMML samples. JMML samples show a statistically significant increase in methylation as compared to HG (P < 0.0001).
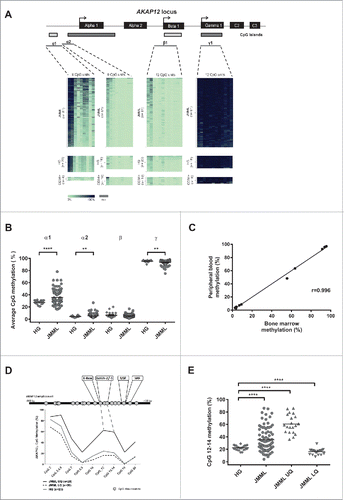
Figure 2. AKAP12α expression and promoter methylation. (A) AKAP12α mRNA expression levels measured by qRT-PCR in five healthy granulocyte samples and 19 JMML patient samples are shown. Horizontal lines represent mean values. The AKAP12α mRNA expression was significantly lower in JMML patients as compared to HG (P = 0.0002; Mann-Whitney U test). (B) Schematic representation of the AKAP12α promoter sequence with and without GATA-motif cloned into the CpG-free (pCpGL) luciferase reporter vector. K562 cells were transfected either with an unmethylated or with an in vitro methylated pCpGL vector construct. Luciferase activity was measured and normalized to the unmethylated Renilla-luciferase construct. The results are relative to the activity of the unmethylated AKAP12α promoter. The bar chart shows the mean luciferase activity ± SD (n = 4) of the methylated promoter construct and the unmethylated construct. Statistical significance was analyzed by a Student's t-test.
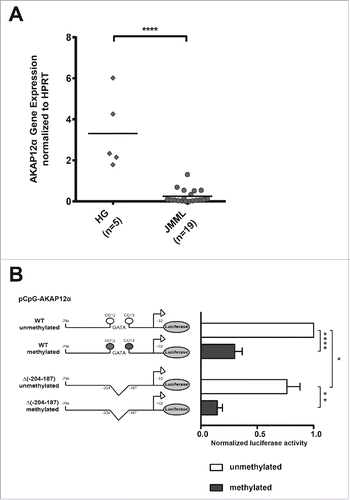
Figure 3. Epigenetic silencing of AKAP12α in RAS-mutated AML cell lines can be reverted by DAC treatment. Time course of AKAP12α1 promoter methylation in Kasumi-1 (A), and HL-60 cells (B) upon treatment with either SB939, DAC or a combination of both drugs and subsequent drug withdrawal. Cells were treated with the respective drug(s) for 72 h (day 1–3) and then observed for 11 d upon drug withdrawal (day 6–14). Time course of AKAP12α mRNA expression in Kasumi-1 (C), and HL-60 cells (D) upon drug treatment and subsequent drug withdrawal. Untreated control cells (Ctrl) are shown as light gray bars, time-points under drug treatment are shown in dark gray, and time-points upon drug withdrawal are depicted as black bars.
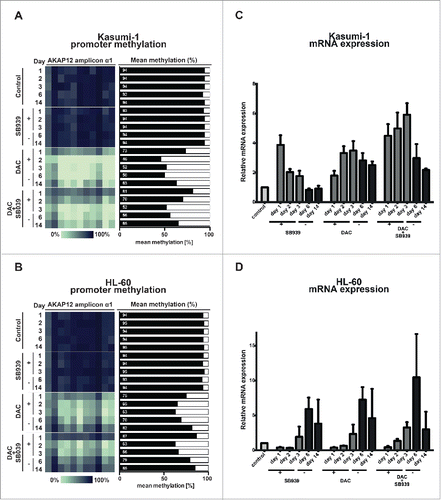
Table 1 Correlation of AKAP12 promoter methylation with clinical parameters in JMML patients (N = 81).