Figures & data
Table 1. Characteristics of mother-newborn pairs.
Table 2. Characteristics of particulate air pollution exposure. Averaged for each mother-newborn pair during the different time windows during pregnancy.
Figure 1. Associations of relative miRNA expression with in utero exposure to air pollution. Associations are presented as percentage changes in relative miRNA (miR-16, miR-20a, miR-21, miR-34a, miR-146a, and miR-222) expression across the three trimesters of pregnancy, for each 5 µg/m3 increase in PM2.5 exposure (black square) and in NO2 exposure (grey triangle). Estimates were adjusted for newborn's gender, gestational age (weeks) and ethnicity (European, non-European), maternal age (years), pre-gestational BMI (kg/m2), smoking status (never-, past- or current-smoker), educational status (low, middle or high), parity (1, 2, or ≥3), seasonality at conception and apparent temperature (during the third trimester). Asterisk (*) indicates statistically significant (P < 0.05).
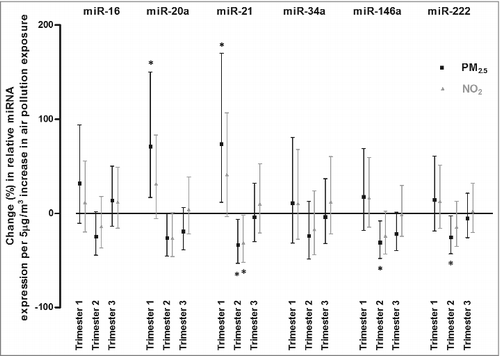
Table 3. In silico putative mRNA targets for placental miRNAs under study. For each miRNA, the mRNA targets (n = 15), description, function and the experimentally validated methods are indicated.
Table 4. Functional enrichment analysis for the putative target genes of deregulated miRNAs in association with air pollution exposure (miR-20a, miR-21, miR-146a, and miR-222). The top 5 enriched MetaCore™ pathways, the P-value, FDR (P-value corrected for multiple testing) and the genes present in our dataset and involved in the listed pathways are provided.
Figure 2. Common putative pathways regulated by identified targets of the significant miRNAs. The top 10 shared pathways for targets of miR-20a, miR-21, miR-146a, and miR-222 are ranked based on their minimum P-value, provided by MetaCore™. Pathways regulated by miR-20a are indicated with orange bars, miR-21 with blue bars, miR-146a with red bars, and miR-222 with green bars. Size of the bars is indicative of the P-value for that respective miRNA.
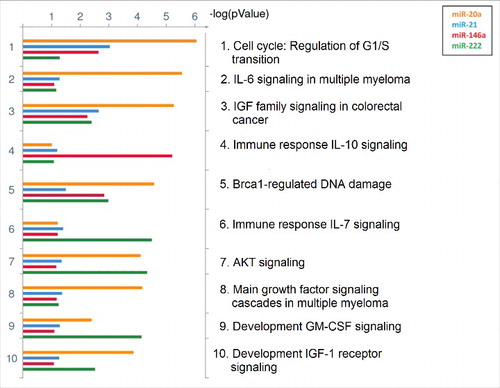
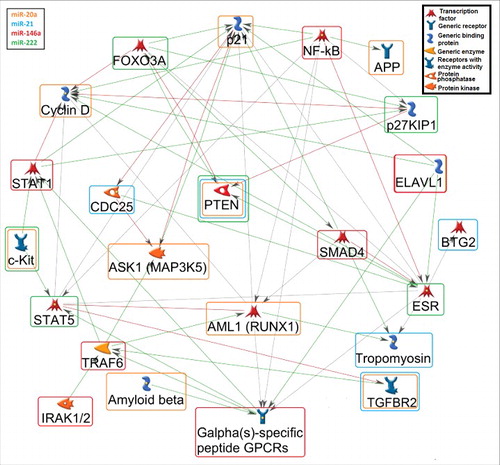
Figure 3. Gene network among the putative miRNA targets. A gene network (MetaCore™) was generated for the potential connections of at least two miRNA-targets. The orange rounded rectangle corresponds to miR-20a, blue to miR-21, red to miR-146a, and green to miR-222 targets. The green arrows show activation, the red arrows indicate inhibition, and the grey arrows are unspecified connections. Details for the genes shown in the figure: AML1 (RUNX1): Runt-related transcription factor 1; APP: Amyloid beta (A4) precursor protein; ASK1 (MAP3K5): Mitogen-Activated Protein Kinase Kinase Kinase 5; BTG2: BTG family, member 2; CCND: Cyclin D; CDC25: Cell Division Cycle 25; CDKN1A (p21): Cyclin-Dependent Kinase Inhibitor 1A; CDKN1B (p21KIP1): Cyclin-dependent kinase inhibitor 1A/B; ELAVL1: ELAV Like RNA Binding Protein 1; ESR: Estrogen receptor; FOXO3A Forkhead box O 3A; GPCRs (CXCR4): Chemokine receptor 4 (G Protein-Coupled Receptors); IRAK1/2: Interleukin-1 Receptor-Associated Kinase 1/2; c-KIT: v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog; NFKB: Nuclear factor of kappa light polypeptide gene enhancer in B-cells; PTEN: Phosphatase and tensin homolog; SMAD4: SMAD family member 4; STAT1/5: Signal Transducer And Activator Of Transcription 1/5; TGFBR2: Transforming growth factor, beta receptor II; TPM1: Tropomyosin; TRAF6: TNF receptor-associated factor 6.