Figures & data
Figure 1. 450K methylation array and pyrosequencing data. Box plots showing miR26A1 methylation levels for CpG site cg26054057 in IGHV-mutated (n = 9) and IGHV-unmutated (n = 9) CLL (A) and MCL (B) samples based on 450K methylation array data. Box plots showing percentage of DNA methylation levels of miR26A1, as assessed by pyrosequencing in CLL (C) and MCL (D) primary samples along with normal B-cell controls.
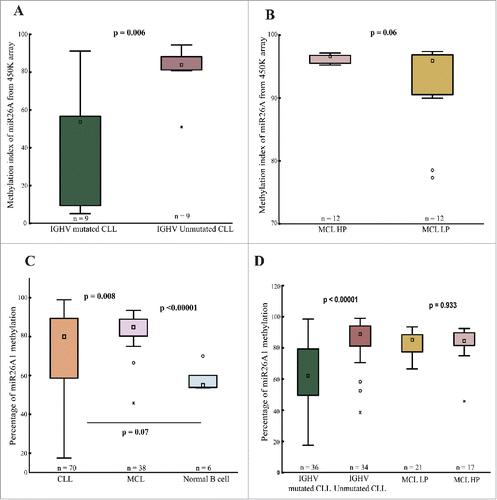
Figure 2. miR26A1 DNA methylation levels predicts overall survival in CLL. Kaplan-Meier curves for CLL patients with high and low DNA methylation (A). Scatter plots showing methylation levels of miR26A1 plotted against survival data in years (B). Black dots represent IGHV-mutated samples and red dots IGHV-unmutated samples. This scatter plot is again divided into 4 quartiles based on the median survival time in CLL (which is 10 years) and the median methylation percentage levels (which is 80% based on the pyrosequencing data).
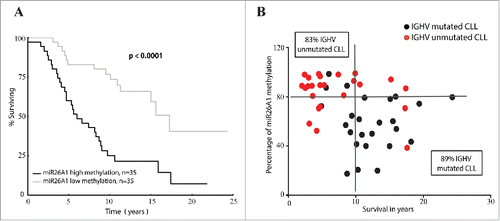
Figure 3. Correlation between miR26A1 and EZH2 expression. Scatter plot showing inverse correlation between EZH2 and miR26A1 expression levels in CLL patient samples (P < 0.05 and r = −0.4484) (A). Relative expression levels of miR26A1 in negative control mimic miRNA and miR26A1 mimic miRNA transfected cell lines. Left panel shows data in HG3 and MEC1 CLL cell lines and right panel the corresponding data in Z138 and GRANTA519 MCL cell lines (B). Western blot analysis of EZH2 expression levels in miR26A1 and control mimic miRNA transfected HG3/MEC1 CLL cell lines (left panel) and Z138/GRANTA519 MCL cell lines (right panel). GAPDH was used as internal loading controls. Histograms below Western blots show calculated band intensities based on 2 independent transfections (C). Percentage of EZH2 protein expression using FACS analysis in CLL (left) and MCL (right) cell lines (D).
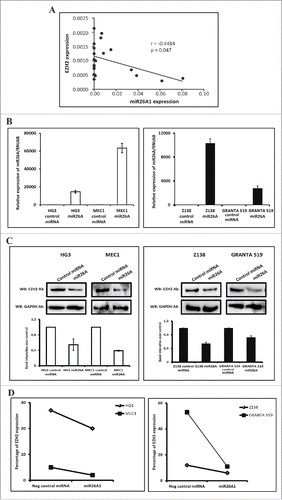
Figure 4. Effect on miR26A1 and EZH2 expression levels after DAC treatment using CLL and MCL cell lines. Relative expression levels of miR26A1 in Z138 and GRANTA519 cell lines (left side) and HG3 and MEC1 cell lines (right side) after DAC treatment using increasing concentrations (A-B). Relative EZH2 mRNA expression for the same MCL (left side) and CLL (right side) DAC treated cell lines (C-D). All samples were treated for 3 d; DAC was changed every 24 h.
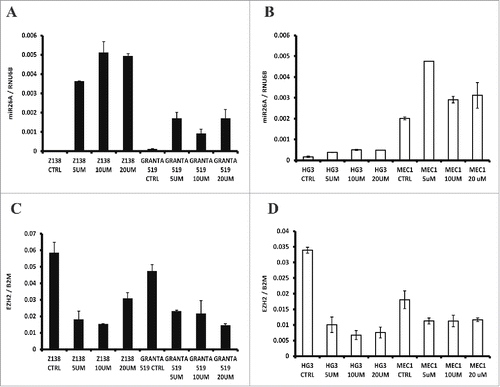
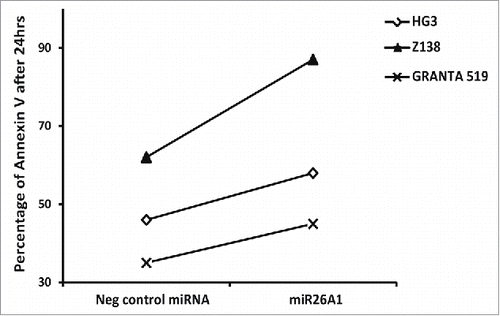
Figure 5. Overexpression of miR26A1 induces apoptosis in CLL and MCL cell lines. The percentage of Annexin V-positive cells analyzed using FACS for control and miR26A1 mimic miRNA transfected cell lines. The percentage of Annexin V positive cells show the average of 2 independent sets. The Annexin V levels were measured after 24, 48, and 72 h.