Figures & data
Figure 1. Gene organization and protein structure of the mussel macroH2A. A) Gene organization of human (Hs) and mussel (Mg) macroH2A genes (represented at an arbitrary scale for comparison purposes). Hs macroH2A.1.1 and Hs macroH2A.1.2 are splicing variants from the Hs macroH2A.1 gene. The length of exons and introns (number of nucleotides) is indicated at the corresponding positions (mE, mussel exon; hE, human exon). Exon numbering in humans was assigned after.Citation6 Red open boxes at 5′ and 3′ positions represent untranslated regions (UTRs), indicating their length in nucleotides. B) Secondary structure prediction for different macroH2A variants from metazoan animals including vertebrates and invertebrates. Red boxes and red arrows indicate the presence of α-helices and β-sheets, respectively, at the amino acid positions indicated. C) Predicted tertiary structure for Mg macroH2A [modeled using Phyre2 Citation73] compared with those of human macroH2A proteins.
![Figure 1. Gene organization and protein structure of the mussel macroH2A. A) Gene organization of human (Hs) and mussel (Mg) macroH2A genes (represented at an arbitrary scale for comparison purposes). Hs macroH2A.1.1 and Hs macroH2A.1.2 are splicing variants from the Hs macroH2A.1 gene. The length of exons and introns (number of nucleotides) is indicated at the corresponding positions (mE, mussel exon; hE, human exon). Exon numbering in humans was assigned after.Citation6 Red open boxes at 5′ and 3′ positions represent untranslated regions (UTRs), indicating their length in nucleotides. B) Secondary structure prediction for different macroH2A variants from metazoan animals including vertebrates and invertebrates. Red boxes and red arrows indicate the presence of α-helices and β-sheets, respectively, at the amino acid positions indicated. C) Predicted tertiary structure for Mg macroH2A [modeled using Phyre2 Citation73] compared with those of human macroH2A proteins.](/cms/asset/d177b22a-913d-4dbb-9cd8-31f65d7a71fd/kepi_a_1172161_f0001_oc.gif)
Figure 2. New evolutionary context for macroH2A. Schematic macroH2A protein phylogeny (see complete tree in Fig. S3) illustrating the wide distribution of this histone variant across metazoans and in the holozoan Capsaspora. The progressive specialization of macroH2A is evident in vertebrates, resulting in the differentiation of macroH2A.1 (purple) and macroH2A.2 (pink), different from macroH2A from non-vertebrates (green). The taxonomic classification of the organisms represented in the tree is indicated in the right margin of the figure. The tree was rooted with the human histone H2A.Z, as it constitutes a sister monophyletic group of macroH2A within the H2A phylogeny.Citation34,38
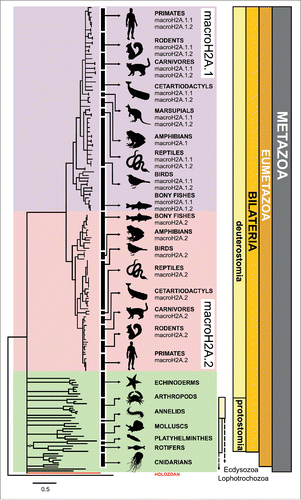
Figure 3. Detection of the macroH2A protein detection in non-vertebrates. Upper gel, SDS-PAGE of HCl-extracted histones from representative organisms including: mussel (M. californianus) hepatopancreas (lane 1), tick (A. maculatum) salivary glands (lane 2), sea urchin (S. purpuratus) male gonad (lane 3) and amphioxus (B. floridae, lane 4). CM, chicken erythrocyte histones used as marker. Lower gel, western blot analysis of the HCl-extracted proteins using the invertebrate-specific anti-macroH2A antibody (M12) developed in the present work. Mussel recombinant macroH2A (rM) was used as positive control, and anti-H4 antibody for sample normalization.
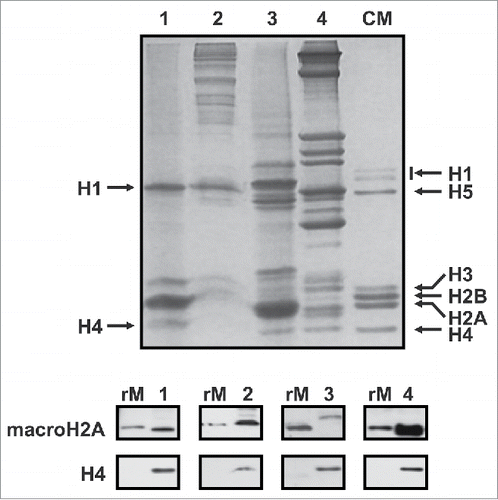
Figure 4. Tissue and chromatin-specific distribution of mussel macroH2A. A) Upper gel, SDS-PAGE of HCl-extracted proteins from different somatic tissues of the mussel M. californianus including: hepatopancreas (lane 1), muscle (lane 2), gills (lane 3) and hemolymph (lane 4). Lower gel, protein gel blot analysis of the HCl-extracted proteins using the invertebrate-specific anti-macroH2A antibody (M12) developed in the present work. B) Upper gel, SDS-PAGE of HCl-extracted proteins from different germinal tissues of the mussel M. californianus including: sperm (lane 1), male gonad (lane 2) and female gonad (lane 3). Lower gel, western blot analysis of the HCl-extracted proteins as in A). CM, chicken erythrocyte histones used as marker; rM, mussel recombinant macroH2A used as positive control; H4, anti-H4 antibodies used for sample normalization.
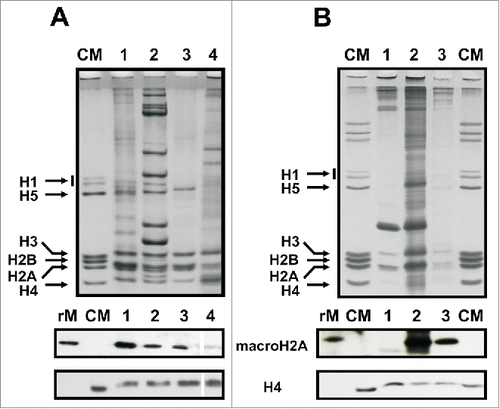
Figure 5. MacroH2A distribution in the chromatin of vertebrate and non-vertebrate organisms. A) Upper gel, SDS-PAGE analysis of the nuclear (N), SI, SE and P chromatin fractions obtained after digestion of mussel hepatopancreas nuclei with micrococcal nuclease. Lower gel, protein gel blot analysis of the HCl-extracted proteins as in A). B) Upper gel, SDS-PAGE analysis of the nuclear (N), SI, SE and P chromatin fractions obtained after digestion of mouse liver nuclei with micrococcal nuclease. Lower gel, Western blot analysis of the HCl-extracted proteins incubated with mouse-specific anti-macroH2A.1.2 and anti-macroH2A.2 antibodies (sequentially). CM, chicken erythrocyte histones used as marker; rM, mussel recombinant macroH2A used as positive control; H3 and H4, anti-H3 and anti-H4 antibodies used for sample normalization.
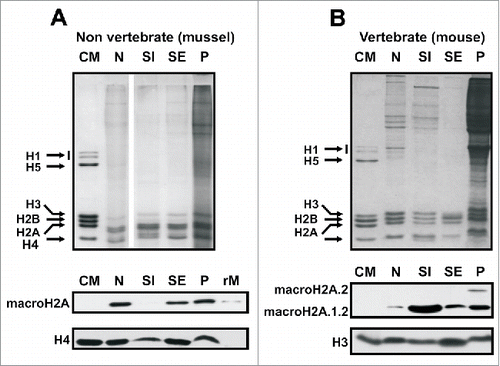
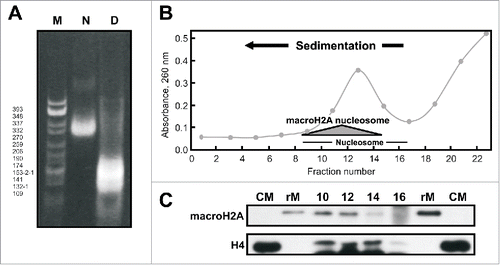
Figure 6. MacroH2A-nucleosomes in the mussel Mytilus. A) Native 4% (w/v) PAGE of the SI chromatin fraction obtained from M. californianus hepatopancreas. N, nucleosome; D, DNA; M, CfoI-digested pBR322 plasmid DNA used as marker. B) Sucrose gradient fractionation of the SI fraction obtained from M. californianus hepatopancreas. The shaded triangle corresponds to the position of the nucleosomes containing macroH2A. C) Western blot analysis of the gradient fractions 10–16 using the invertebrate-specific anti-macroH2A antibody (M12) developed in the present work and anti-H4 antibody. Mussel recombinant macroH2A (rM) was used as positive control. CM, chicken erythrocyte histones used as marker.