Figures & data
Figure 1. Flowchart for Experimental and Analysis Pipeline. Both humans and mice underwent MethylPlex enrichment prior to sequencing. Reads were Repeat Masked and used to identify group level differences, and aligned to reference genomes to identify individual differences in repeat DNA methylation. Asterisks denote significance, *P < 0.05, **P < 0.01.
Figure 2. Percent CpG Enrichment by Exposure. Both human and mouse samples exhibited increased CpG content after MethylPlex enrichment. (A) The human low BPA group was slightly but significantly lower than the medium group (P = 0.01), and the high group (P < 0.05). (B) The mouse samples exhibited no significant differences in CpG enrichment by exposure group.
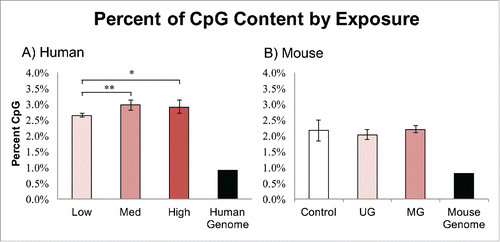
Figure 3. Percent Reads by Repeat Class and Exposure. (A) A subset of classes in human was hypomethylated in the medium BPA group. (B) No classes were differentially methylated by exposure group in mice. Asterisks denote significance, *P < 0.05, **P < 0.01.
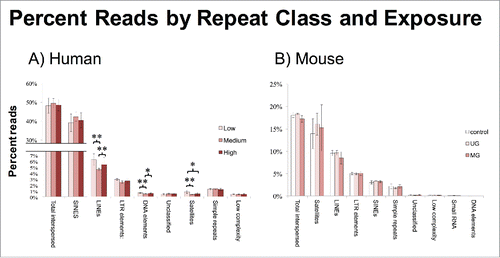
Figure 4. Percent Reads by Repeat Family and Exposure. (A) A subset of repeat families was hypomethylated in the medium BPA group. (B) No families were differentially methylated by exposure group in mice. Asterisks denote significance, *P < 0.05, **P < 0.01.
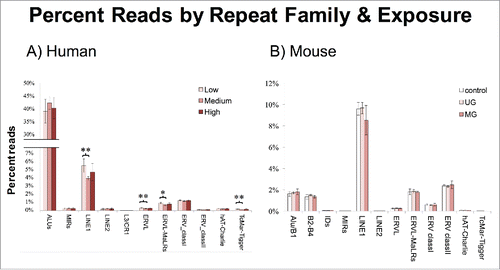
Table 1. Repeat subfamilies differentially methylated by exposure.
Figure 5. Individual Transposon Detection Pipeline. Both mouse and human reads were filtered for reads containing partial unique and partial repetitive sequence. The unique sequence was mapped to the reference genome and read count differences corresponding to DNA methylation differences were tested for significance with EdgeR.
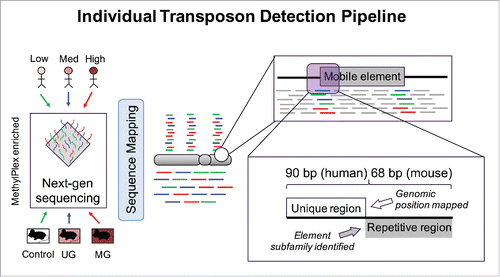
Figure 6. Individual Repeats Differentially Methylated by Exposure. (A) Number of human transposon insertions exhibiting significantly different read counts across BPA exposure groups. (B) Mouse transposon insertions exhibiting differential read counts across BPA treatment groups.
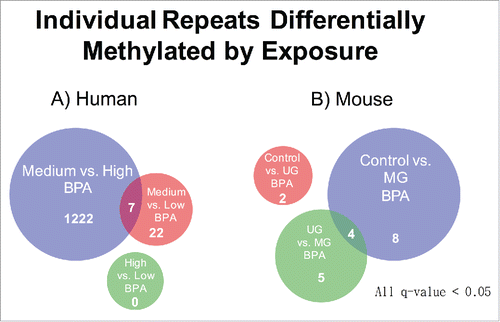
Table 2. Genomic distribution counts of differentially methylated transposons.
