Figures & data
Figure 1. Strategy to identify epigenetically downregulated genes in pN+ OSCC. On the left: published gene signatures predictive of pN-status in OSCC were used to identified significantly downregulated genes in pN+ OSCC.Citation20-22 On the right: MethylCap-Seq was performed on 6 pN0 OSCC and pN+ OSCC.Citation18 All reads of MCs in gene promoter regions were ranked according to the likelihood of differential methylation and an approximate FDR. The 5,000 MCs with the lowest FDR were further tested by Mann-Whitney-U. The MC associated with genes without annotated gene functions were excluded. In the middle: the gene signature and methylation data were compared to select epigenetically regulated genes in pN+ OSCC (n = 23). From these 23 genes, epigenetically downregulated genes in pN+ OSCC were selected. Based on the amount of mRNA downregulation, statistical differences in methylation between pN0 and pN+ OSCC, and positive and negative predictive value, RAB25 was selected as the most significantly epigenetically downregulated gene in pN+ OSCC compared to pN0 OSCC.
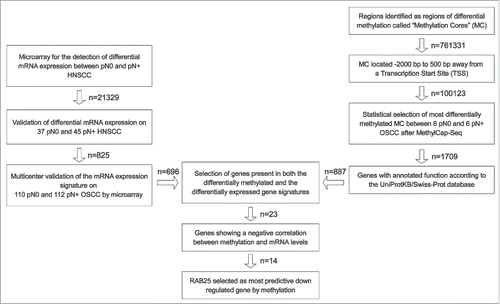
Table 1. Epigenetically downregulated genes in pN+ OSCC. All 14 potentially epigenetically downregulated genes in pN+ OSCC compared to pN0 OSCC after cross-reference of expression microarray and MethylCap-Seq data (see ). The positive and negative predictive value of the reads for pN+ status, associated hypermethylation, read distribution between pN0 and pN+ OSCC, and predictive value of the methylation data are illustrated. P-value for the differential DNA methylation was calculated using the Mann-Whitney-U test. Positive and negative predictive value for the methylation status of all MCs were calculated as follows: OOSCC with a read count of ≥ 3 reads were considered true positives and OOSCC with a count read<3 were considered true negatives. Subsequently, the positive predictive value was then calculated as: (true positive pN+ OOSCC) / (true positive pN+ OOSCC + false positive pN0 OOSCC). Finally, the negative predictive value was calculated as: (true negative pN0 OOSCC) / (true negative pN0 OOSCC + false negative pN+ OOSCC).
Figure 2. RAB25 mRNA levels in relation with the 3 RAB25 TSS 450K probes (cg09243900, cg15896939, and cg19580810) methylation levels in the TCGA OSCC cohort. (A) RAB25 methylation levels compared between OSCC with high RAB25 mRNA levels and OSCC with low RAB25 mRNA levels. The M-values of the 3 RAB25 Infinium 450K promoter probes were significantly higher in OSCC with low RAB25 mRNA z-scores compared to OSCC with high RAB25 mRNA z-scores. (B) Spearman correlations between RAB25 methylation and RAB25 mRNA levels. All 3 RAB25 promoter probes showed a significant negative correlation between RAB25 promoter probe M-values and RAB25 mRNA z-scores.
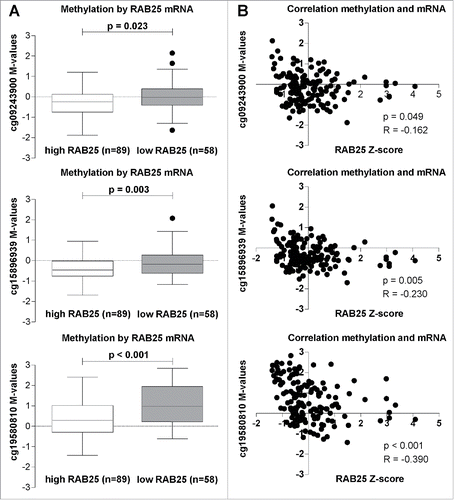
Figure 3. RAB25 expression levels between pN0 and pN+ OSCC in the UMCG and TCGA OSCC cohort. (A) pN+ OSCC in the TCGA cohort (n = 86) express significantly less RAB25 mRNA than pN0 OSCC (n = 61), as revealed by Mann-Whitney-U test. (B) pN + OSCC in the UMCG cohort (n = 87) have significantly less RAB25-positive tumor cells than pN0 OSCC (n = 91), as revealed by Mann-Whitney-U test.
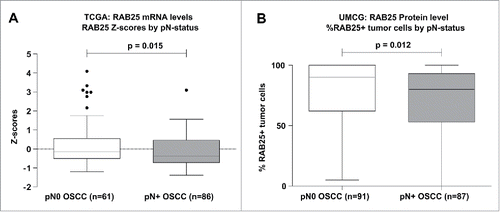
Table 2. Correlations between RAB25 expression and tumor characteristics. A) Associations between RAB25 mRNA expression and the clinical characteristics of the TCGA OSCC cohort. B) Associations between RAB25 protein expression and the clinical characteristics of the UMCG OSCC cohort.
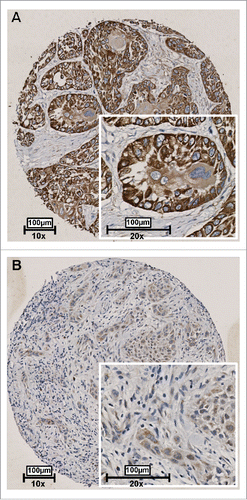
Figure 4. Representative examples of RAB25 expression in 2 OSCC as detected by immunohistochemistry. Tissues were scored by the amount of RAB25-positive cells. (A) Example of a well-differentiated OSCC with a high amount of RAB25-expressing cells. (B) Example of a poorly differentiated OSCC with a very low amount of RAB25-positive cells.