Figures & data
Figure 1. Schematic diagram of the experimental and analytical procedures. Sprague-Dawley (SD) rats were neonatally [postnatal day (PND) 1, 3, 5] treated with 17β-estradiol-3-benzoate (EB) at 25 μg/pup or 2,500 µg/kg body weight (BW), bisphenol A (BPA) at 0.1 µg/pup or 10 µg/kg BW, or corn oil as a control (Ctrl). DNA extracted from PND90 dorsal prostate tissue was subjected to promoter methylation array analysis, and genes with differentially methylated regions and inverse expression correlation were identified and validated.
![Figure 1. Schematic diagram of the experimental and analytical procedures. Sprague-Dawley (SD) rats were neonatally [postnatal day (PND) 1, 3, 5] treated with 17β-estradiol-3-benzoate (EB) at 25 μg/pup or 2,500 µg/kg body weight (BW), bisphenol A (BPA) at 0.1 µg/pup or 10 µg/kg BW, or corn oil as a control (Ctrl). DNA extracted from PND90 dorsal prostate tissue was subjected to promoter methylation array analysis, and genes with differentially methylated regions and inverse expression correlation were identified and validated.](/cms/asset/3aaeca16-9043-45ff-97a8-1f4b2d708db0/kepi_a_1208891_f0001_c.gif)
Table 1. Top networks and bio-functions of the genes associated with neonatal 17β-estradiol-3-benzoate (EB) or bisphenol A (BPA) exposure in postnatal day (PND)90 prostate.
Figure 2. Representative results from genome-wide methylation study. A) Predicted CpG islands (light blue shaded areas) in the promoter region of differentially methylated genes (Pitx3, Wnt10b, Paqr4, Sox2, Chst14, Tpd52, and Creb3l4) identified in this study. TSS stands for transcriptional start site whereas ATG stands for translational start codon. Individual CpG sites are represented by red vertical lines. Dark blue horizontal line marks the region selected for BS-sequencing; B) Position of the BS-sequenced region (blue line) relative to the NimbleGen probes covered regions (dirty yellow lines) of each gene; C) Significant methylation level of each gene in vehicle- (control), EB- and BPA-treated groups, which were measured by NimbleGen array probes (mBar values). The height of the mBar represents the probe intensity; red and green bars represent positive and negative methylation value, respectively, relative to their respective input control.
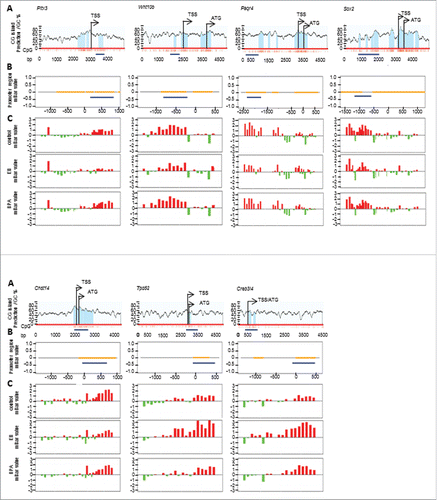
Table 2. Validation of promoter methylation status and gene expression in postnatal day (PND)90 prostate.
Figure 3. Effect of neonatal exposure to EB or BPA on promoter methylation and gene expression in PND90 dorsal prostate. Promoter methylation status (left panel: Scatter Plot) and expression (right panel: Bar Graph) of candidate genes, in PND90 prostate tissues from SD rats treated with either corn oil (Ctrl; white), EB (green), or BPA (red), were analyzed using bisulfite sequencing and qPCR, respectively. Each circle in the scatter plot represents mean ±SEM of methylation percentage (averaged from 6 individual samples/animals) at a single CpG site in the gene promoter region. The % methylation of each CpG site in each sample was determined from bisulfite sequencing data derived from 8–12 clones. Gene expression data were expressed as mean ± SEM from 3 individual samples. Statistical significance was determined by one-way ANOVA and Tukey test when compared to Ctrl. *P < 0.05, **P < 0.01, and ***P < 0.001.
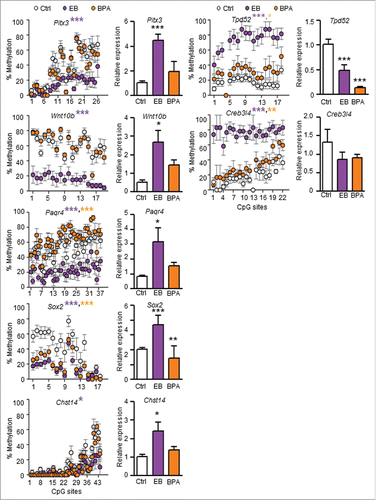
Figure 4. Signaling pathways associated with differentially methylated candidate genes were predicted by Ingenuity Pathway Analysis™. Genes with promoter hypermethylation are shown in red and genes with promoter hypomethylation are shown in green, with color intensity signifying the magnitude of differential methylation. Gray arrows indicate predicted association pathways; purple arrows indicate reported directional pathways.
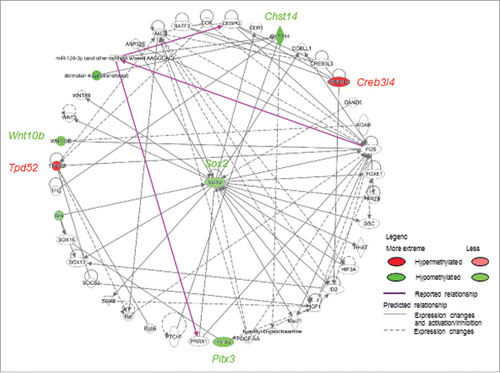
Table 3. Reported bio-function(s) and PCa associations of the seven identified candidate genes with DMRs inversely correlated with gene expression as a result of neonatal 17β-estradiol-3-benzoate/bisphenol A (EB/BPA) exposure.
Figure 5. Effect of 5-aza-cytidine treatment on gene expression in NbE-1 and AIT cells. Gene expression was analyzed by qPCR in rat (A) normal prostate epithelial NbE-1 cells and (B) prostate cancer AIT cells treated with DMSO (Ctrl), or 0.5 µM or 1 µM 5-aza-2-deoxycytidine (5-aza), a DNA methylation inhibitor, for 8 d. Data (mean ± SEM ) is normalized to corresponding Rpl19 levels, and is expressed as fold change vs. Ctrl. *P < 0.05, and **P < 0.001 by one-way ANOVA and Tukey test when compared to Ctrl.
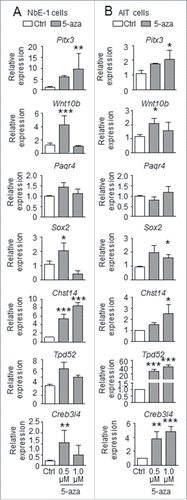
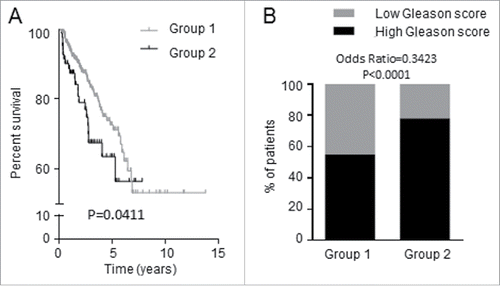
Figure 6. Expression of the 7 candidate genes was associated with shorter recurrence-free survival of PCa patients. TCGA data consisting of 497 PCa patients were dichotomized into Groups 1 and 2 by K-means clustering analysis based on the 7 candidate genes. (A) Group 1 patients have longer time to recurrence than Group 2 patients; the two groups differ in recurrence-free survival. (B) Proportion of PCa patients having tumors with high (≥7) and low (<7) Gleason score.