Figures & data
Table 1. Study subjects.
Figure 1. Differential methylation results. (A) Manhattan plot of the results from analysis of differential methylation by COPD affection status. The blue line represents the threshold for an FDR of 5%. (B) Volcano plot of these results (FDR < 5% and >5% β-diff in blue). Sites with FDR < 5% and β-diff < 5% are shown (cg10800464, cg11635101:MAD1L1, cg07068735:RAB8B, cg00622552:ODF3L1, and cg20802051:AK3L1).
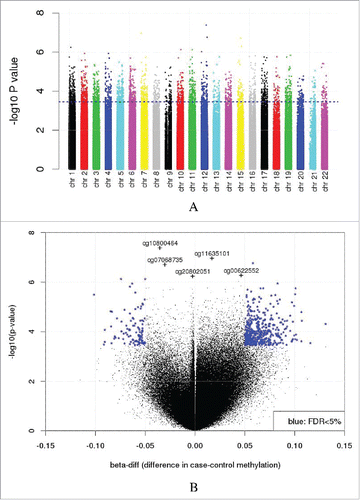
Figure 2. Case and control methylation levels for the top results. Case-control methylation plots (using beanplot R package) for the CpG sites with gene annotations that have at least a 10% β-diff (regression coefficient for affection status). The P-values are adjusted for multiple testing.
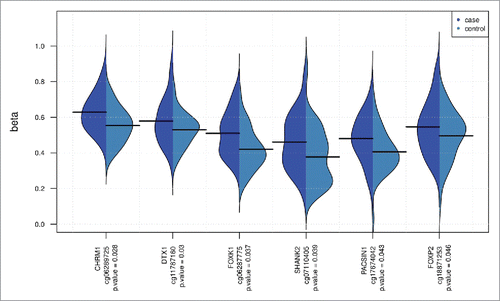
Table 2. Top 10 CpG sites associated with COPD above 5% β-diff threshold, sorted by P-value.
Table 3. CpG sites associated with COPD above 10% β-diff threshold, sorted by P-value.
Figure 3. Case and control methylation and expression levels for 2 multi-site genes. Multi-site results for ESM1 and PITPNM1. Plots of COPD case-control methylation for CpG sites with 5% FDR and β-diff > 1%; ESM1 (A), PITPNM1 (B). Corresponding gene expression plots for these genes (case-control differential expression P-values are unadjusted); ESM1 (C), PITPNM1 (D).
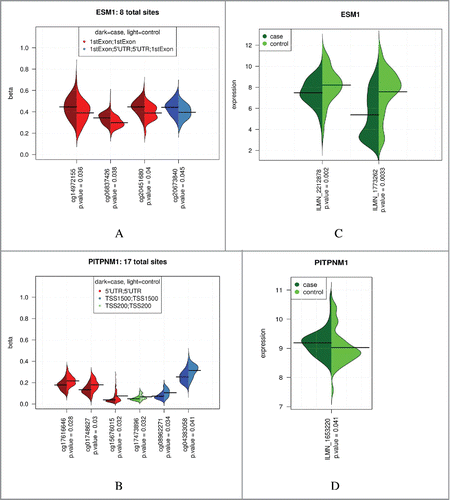
Table 4. Genes most enriched (hypergeometric test P <0.25) with differentially methylated CpG sites that are associated with COPD affection status (multi-site genes).
Table 5. Intersection by gene annotation of the sub-genome-wide significant (P-value < 0.01) COPD GWAS results and the differential methylation results (5% FDR and β-diff > 5%); top ten results sorted by differential methylation P-value.
