Figures & data
Table 1. Characteristics of study participants (n = 20) classified by body mass index.
Figure 1. All methylation sites detected in our whole-blood samples using reduced representation bisulfite sequencing technology were mapped (a) in the context of gene regions and (b) CpG island features. Regions were defined using UCSC browser RefGene and CpG island tracks (see methods). The promoter region was defined as 1000 bp upstream of the transcription start site (TSS); CpG island is a 200–3000 bp stretch of DNA with a C+G content of 50% and observed CpG/expected CpG exceeding 0.6; North (N) and South (S) shores flank the CpG island by 0–2000 bp; the North (N) and South (S) shelf flank the shores by 2000 bp (2000–4000 bp from the island). UTR, untranslated region.
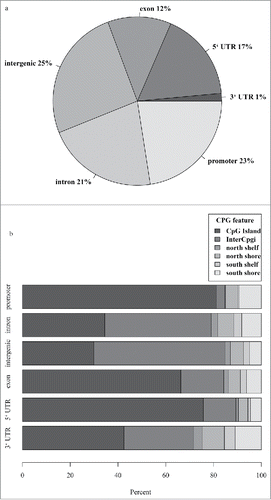
Table 2. Whole-blood differentially methylated cytosines (DMCs; q < 0.05) between lean and obese groupings.
Table 3. Regression analysis of the differentially methylated cytosines (DMCs; q < 0.05) predicted by M value after adjusting for age and sex.
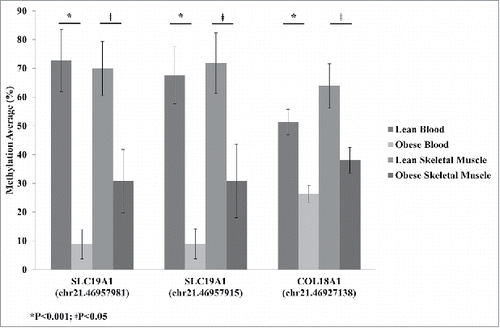
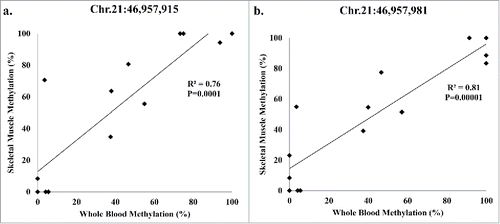
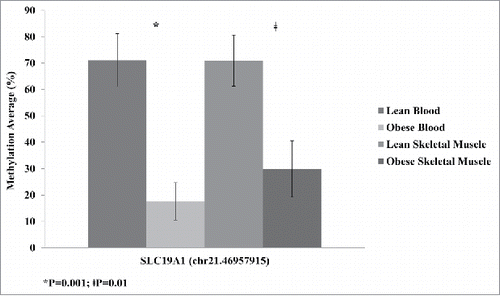
Figure 2. Average methylation detected by reduced representation bisulfite sequencing (RRBS) for SLC19A1 sites Chr.21:46,957,981 and Chr.21:46,957,915 and COL18A1 site Chr.21:46,927,138 for lean and obese in both blood and skeletal muscle. Significance based on independent sample t-tests.