Figures & data
Figure 1. Analysis of the DNA binding capacity of GBD-ROS1_CD in vitro. (A) Schematic diagram of the His-tagged GBD-ROS1_CD recombinant fusion protein. ROS1_CD comprises residues 295–1393 of ROS1, and contains a non-contiguous DNA glycosylase domain distributed over two segments (blue and red) separated by a non-structured linker region (striped), and a C-terminal domain (yellow). (B) Increasing concentrations of purified GBD-ROS1_CD were incubated at 25°C for 30 min with 10 nM of fluorescein-labeled DNA duplex containing (UAS) or not (no-UAS) the sequence targeted by GBD. After non-denaturing gel electrophoresis, gels were scanned to detect fluorescein-labeled DNA. Protein–DNA complexes were identified by their retarded mobility compared with that of free DNA. A representative gel for each substrate is shown. (C) Percentage of protein–DNA complexes versus protein concentration. All bands with slower mobility were used in quantitation of bound protein. Values are mean ± SE (error bars) from three independent experiments.
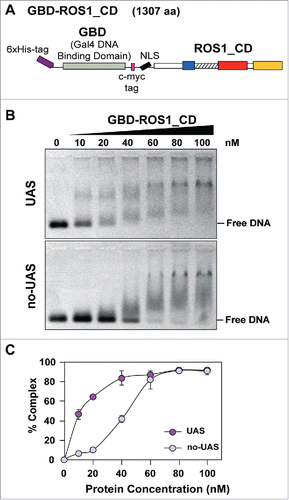
Figure 2. Transient expression of GBD-ROS1_CD and its mutant variants in HEK293 cells. (A-B) Schematic diagrams of effector and reporter constructs used for co-transfection of HEK293 cells. Control effector constructs contain either a mutation in the ROS1 catalytic domain (GBD-ROS1_CDmut) or two mutations in GBD that abolish binding to target UAS sequences (GBDmut-ROS1_CD). Reporter constructs contain the TK promoter fused to the firefly luciferase gene. The targeted version (5xUAS-TK-Luc) includes five copies of GBD binding sites upstream the TK promoter that are absent in the non-targeted control (TK-Luc). (C) Transient expression of GBD-ROS1_CD fusion proteins in HEK293 cells. Western-blot analysis with an anti-Flag antibody was performed in cell extracts (80 μg) prepared 48 h after co-transfection of different effector constructs with either the targeted (upper panels) or the non-targeted (lower panels) methylated reporter plasmid. Actin was used as an input control.
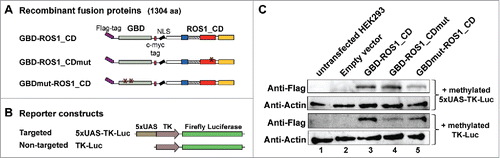
Figure 3. Targeted reactivation of luciferase activity by transient expression of GBD-ROS1_CD. Cells were co-transfected with the indicated effector construct and either the targeted (A) or non-targeted (B) version of the in vitro-methylated reporter plasmid. Luciferase activity, determined 48 h after co-transfection, is shown relative to that detected after co-transfection with empty vector and unmethylated reporter. Values are means ± SE (error bars) from three independent transfection experiments. Asterisks indicate statistically significant differences (P < 0.001; Student's unpaired t-test).
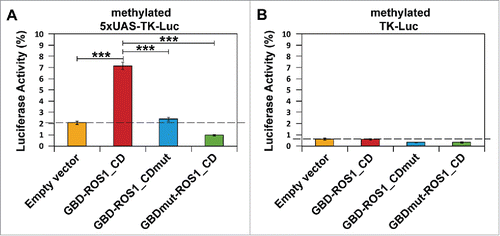
Figure 4. Targeted transcriptional activation of luciferase reporter gene by transient expression of GBD-ROS1. Cells were co-transfected with the indicated effector construct and either the targeted (A) or non-targeted (B) version of the in vitro methylated reporter plasmid. Levels of firefly luciferase transcript, assessed 48 h after transfection by qRT-PCR and normalized to GAPDH, are shown relative to those detected after co-transfection with empty vector and unmethylated reporter. Values are means ± SE (error bars) from three independent transfection experiments. Asterisks indicate statistically significant differences (P < 0.05; Student's unpaired t-test).
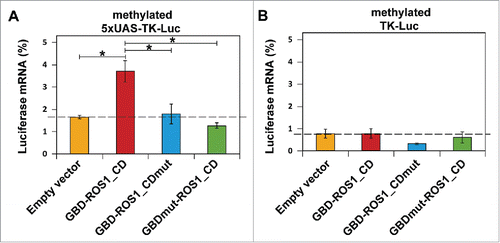
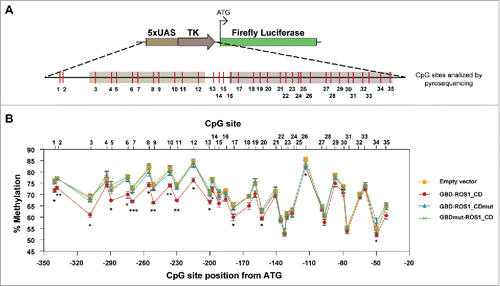
Figure 5. DNA methylation analysis of the targeted 5xUAS-TK region after transient expression of GBD-ROS1_CD. (A) Schematic diagram of the analyzed region. CpG sites are shown in red. (B) Quantitative methylation analysis. Cells were co-transfected with the indicated effector construct and the targeted methylated reporter plasmid. Plasmid DNA was re-isolated 48 h after co-transfection, bisulfite-treated, PCR-amplified, and pyrosequenced. The graph shows the percentage of methylation at different positions. Values are means ± SE (error bars) from three independent transfection experiments. Asterisks indicate statistically significant differences between GBD-ROS1 and the empty vector (*: P < 0.05; **: P < 0.01; ***: P < 0.001; Student's unpaired t-test).