Figures & data
Table 1. Promoter SNP genotype and methylation of MLH1 CpG island and shore in CRC cell lines. A panel of five colorectal carcinoma cell lines was genotyped by Sanger sequencing to determine genotype of rs1800734. MethyLight was utilized to determine the percentage of fully methylated alleles at the CpG island and shore of MLH1. Average PMR (percent methylated reference) of two duplicate reactions is shown.
Figure 1. Bisulfite sequencing of MLH1 CpG island and shore in colorectal cancer cell lines. (A) Three overlapping regions upstream of MLH1 were amplified by bisulfite sequencing in cell lines: Amplicon A, Amplicon B, and Amplicon C. Amplicon A and B overlap at 3 CpGs. Amplicon B and C overlap at 1 CpG. (B) Representative unmethylated clone for each of the three amplicons, located in the CpG shore, middle region, and CpG island. Empty circles represent unmethylated CpG sites and filled in circles represent methylated CpG sites. (C) Methylation patterns in the colorectal cancer cell lines HCT 116, LS 174T, SNU-C2B, COLO 320HSR, and HCT-15 with rs1800734 genotype indicated. Each horizontal line represents a single DNA strand and circles represent individual CpG sites.
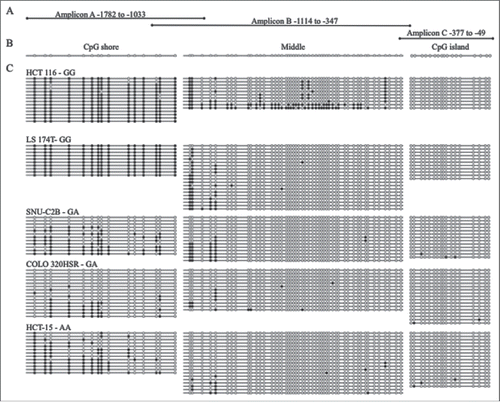
Figure 2. ChIP analysis of AP4 occupancy at the MLH1 CpG island and shore region. (A) Chromatin immunoprecipitation followed by qPCR was performed at five regions upstream of MLH1, located in the CpG shore (S1 and S2), middle region (M1 and M2), and promoter CpG island (P1). MethyLight and bisulfite sequencing regions interrogated are also indicated. SNP location is indicated by gray circle. (B) Experiments were performed in HCT 116 (GG), SNU-C2B (GA), and HCT-15 (AA) cell lines to compare AP4 occupancy among genotypes of SNP rs1800734. Three biological replicates of each cell line were run in triplicate and averaged after ChIP-qPCR. Error bars represent standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001 by independent samples t-test.
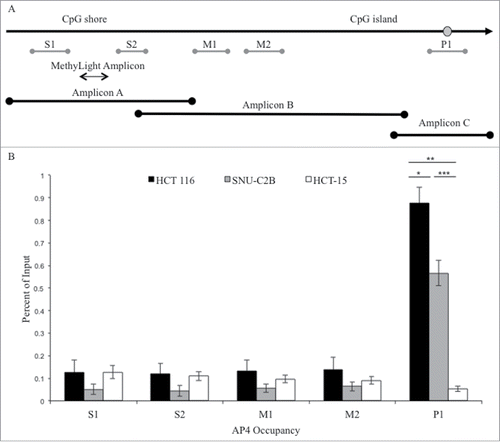
Figure 3. ChIP analysis of histone modifications at the MLH1 CpG island and shore. Chromatin immunoprecipitation was performed in HCT 116 (GG), SNU-C2B (GA), and HCT-15 (AA) cell lines to compare histone modifications among genotypes of SNP rs1800734. Three biological replicates of each cell line were run in triplicate and averaged after ChIP-qPCR at five regions of the MLH1 CpG island and shore: S1, S2, M1, M2, and P1. Histone modifications include: (A) H3K4me1, (B) H3K4me3, (C) H3K27ac, and (D) H3K27me3. Error bars represent standard deviation. *P < 0.05 by independent samples t-test.
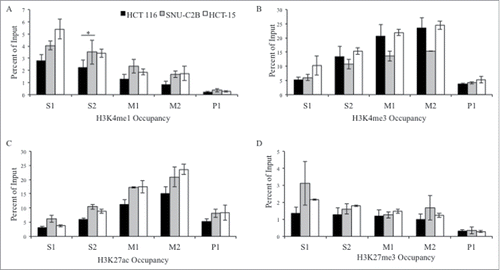
Figure 4. ChIP analysis of Pol II and CTCF at the MLH1 CpG island and shore. Chromatin immunoprecipitation was performed in HCT 116 (GG), SNU-C2B (GA), and HCT-15 (AA) cell lines to compare Pol II and CTCF binding among genotypes of SNP rs1800734. Three biological replicates of each cell line were run in triplicate and averaged after ChIP-qPCR at five regions of the MLH1 CpG island and shore: S1, S2, M1, M2, and P1. (A) Pol II and (B) CTCF were assessed. Error bars represent standard deviation. *P < 0.05 by independent samples t-test.
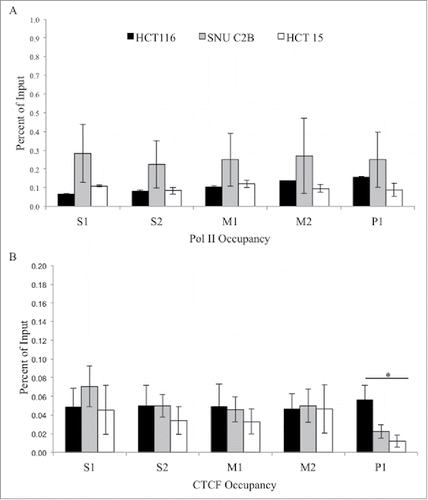
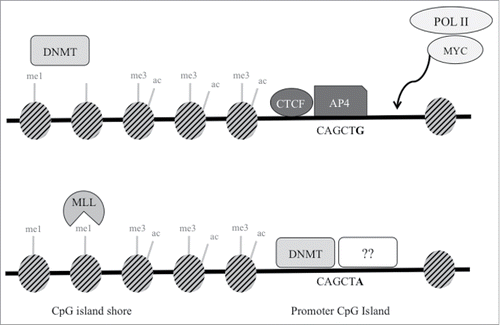
Figure 5. Proposed schematic model of transcription factors and epigenetic regulation at SNP rs1800734. In cells with wild type G allele (top), AP4 transcription factor binds its consensus sequence, potentially interacting with MYC and Pol II to promote transcription of MLH1. Though not demonstrated through experimental results in this study, DNA methyltransferases (DNMT) may maintain DNA methylation at the shore upstream. DNMTs may be prevented from methylating the CpG island in part due to presence of CTCF. In cells with the variant A allele (bottom), AP4 does not bind, which may decrease promoter transcriptional activity. Without the presence of AP4 (or possibly CTCF), DNMTs may methylate the exposed CpG island. This may lead to decreased methylation at the CpG shore and increased H3K4me1, which is deposited by MLL proteins. Other currently unidentified factors may also bind and repress the region further.