Figures & data
Table 1. Epigenetic regulator genes altered in the major forms of human cancer. Genes are listed as their HUGO Gene Nomenclature IDs.
Figure 1. DNA methylation and gene expression consequences after treatment with DNA methylation inhibitors. Circles represent CpG sites, with closed circles indicative of methylated CpGs and open circles representing unmethylated CpGs. Transcription start sites are indicated by the bent arrow. (A) Demethylation of CpG island promoter regions, resulting in gene activity. (B) Gene-body demethylation results in reduced expression. (C) Retained gene silencing by PRC2 occupancy after DNA methylation inhibitor treatment. HMA: Hypomethylating agent.
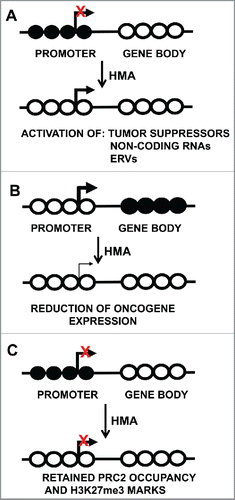
Figure 2. DNA remethylation kinetics after 5-aza-CdR treatment. Top: DNA remethylation of “Fast” (left) and “Slow” (right) CpG sites. Graphs are plotted as percent methylation (y-axis) vs. time after 5-Aza-CdR treatment (x-axis). Bottom: details of Fast and Slow rebounding genes.
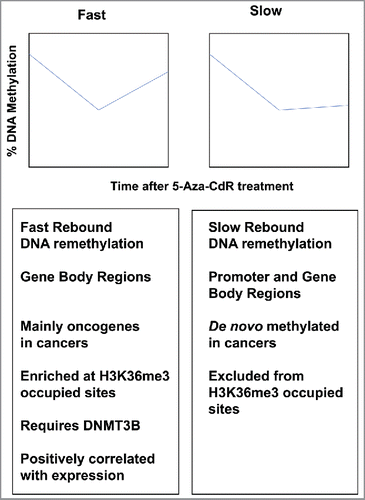
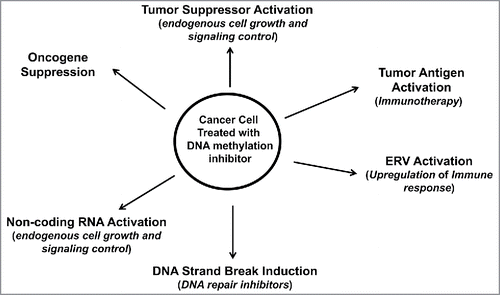
Figure 3. Therapeutic approaches accompanying DNA methylation inhibition for cancer patient treatment. Cancer cells treated with DNA methylation inhibitors have activated tumor suppressors, ERVs and miRNAs, with reduced expression of oncogenes. Activated ERVs promote an immune response that can be further targeted by immune-based therapies. Activated genes may help promote sensitivity to conventional chemotherapies. DNA methylation inhibitors, most notably 5-Aza-CR and 5-Aza-CdR, cause DNA damage, thus sensitizing the treated cell to DNA repair inhibitors.