Figures & data
Figure 1. DNA methylation and splicing in mammals. (A)CTCF Binding to unmethylated CTCF binding sites pause Pol II elongation allowing retention of exon2. (B)CTCF cannot bind to methylated sites resulting in skipping exon2. (C)MeCP2 does not bind to unmethylated sites allowing rapid progression of Pol II resulting in skipping of exon2. (D)MeCP2 binding to methylated site pause elongation of Pol II permitting retention of exon2. Redrawn from Yan et al. 2015.Citation30 CTCF binding site with methylation sensitive CpG in bold: ATGCAGCTAGATGGCGCTC.Citation74
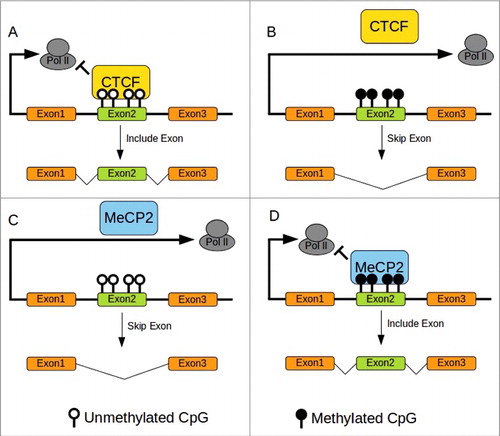
Figure 2. Mechanism of gene imprinting in mammals. (A)Stella binding to H3K9me2 prevent TET3 dependent de-methylation. Maternal (M) DNA is enriched of H3K9me2 compared with paternal (P) and therefore maternal imprinted regions are protected from active de-methylation. (B)After zygote formation and during the firsts cell divisions, level of methylation at imprinted loci is maintained by ZFP57/TRIM28 complex binding to methylated consensus and recruitment of DNMTs. (C)DNMT3a/b interact with H3 tail via ADD domain and their activity is permitted only when H3K4 is unmethylated (H3K4me0). This modification is enriched in the DNA methylated allele of imprinted loci (see text). DNMT3s interact also via a PWWP domain (dotted arrow) to H3K36me3 (enriched in gene bodies of active genes). H3K9me3 methylation is maitained by Setdb1 (continuous arrow). (D)At transcriptionally active imprinted loci the S-phase specific H3.1/H3.2 histones are exchange for the cell cycle independent H3.3 histone. The ATRX/Daxx complex is responsible for the exchange of H3.1/3.2 with H3.3 (dotted arrow). ATRX bind H3K4me0 via an ADD domain and interact also with to H3K9me3. Daxx recruit Setdb1 that maintain H3K9me3 methylation (continuous arrow). Re-draw from Messerschmidt et al. 2014 and Voon and Gibbons 20168,91
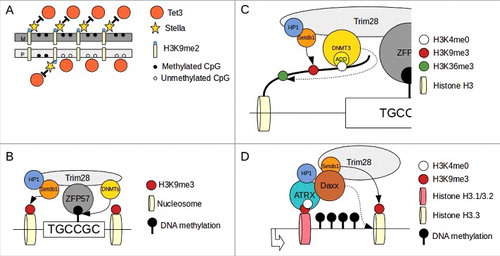
Figure 3. Genetic basis for intragenomic conflict in A. mellifera (left) and B. terrestris (right). In honey bee colonies a single queen (diploid) mates with multiple haploid males. In bumblebee colonies the queen mates only with a single male. Re-drawn from Queller 2003 and Galbraith et al. 2016.Citation4,143
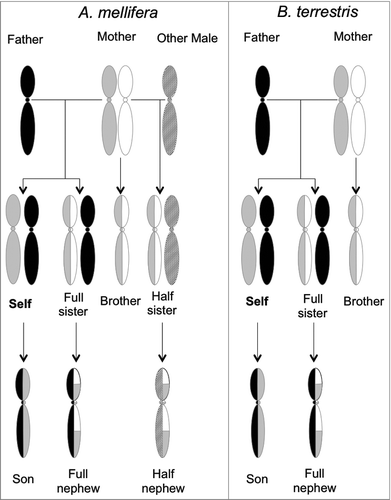
Figure 4. Reciprocal crosses. Females from different lineages (A and B) are crossed reciprocally with single drones of the opposite lineages (B and A). Because of SNPs between the lineages maternal and paternal alleles are recognizable in F1. The global level of expression of maternal (Mat) and paternal (Pat) alleles in a tissue can be tested by RNA-seq. Parental Bias results in an overexpression of the Parental allele (Mat or Pat) in both crosses. Lineage Bias results in an overexpression of the lineage specific allele (A or B). Redraw from Kocher et al 2015.Citation144
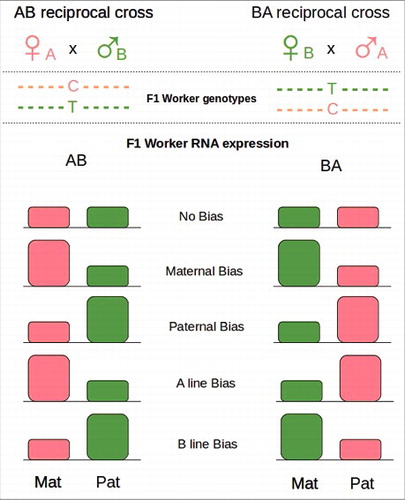
Table 1. Comparison of PSGE in 2 Apis mellifera studies, Kocher et al. 2015 and Galbraith et al. 2016.
Table 2. Useful Technologies for addressing questions of mechanism of GI.
