Figures & data
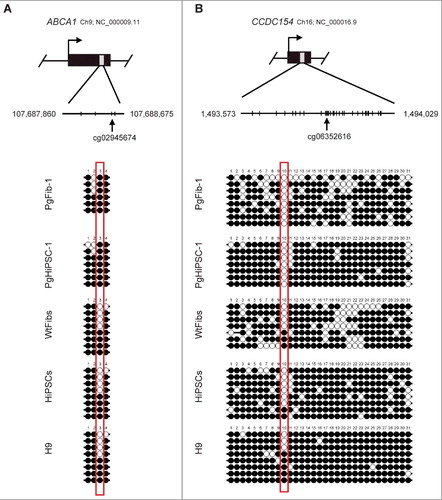
Figure 1. Characterization of parthenogenetic HiPSC lines. (A) Morphology of HiPSCs and parthenogenetic (Pg) HiPSC-1. (B) Immunocytochemistry for pluripotency markers (OCT4 and SSEA4) in biparental HiPSCs and parthenogenetic HiPSC-1. Scale bar = 100 μm. (C) RT-PCR analysis of pluripotency-specific gene expression in WtFibs, PgFib-1, HiPSCs, PgHiPSC-1, and H9. (D) Scatter plots comparing the global gene expression patterns between WtFibs and HiPSCs, PgFib-1 and PgHiPSC-1, H9 and HiPSCs, and H9 and PgHiPSC-1 analyzed by oligonucleotide microarrays. (E) Bisulfite sequencing analysis of OCT4 and NANOG promoter regions in WtFibs HiPSCs, PgFib-1, PgHiPSC-1, and H9. (F) Confirmation of a normal 46, XX karyotype of PgFib-1 and PgHiPSC-1.
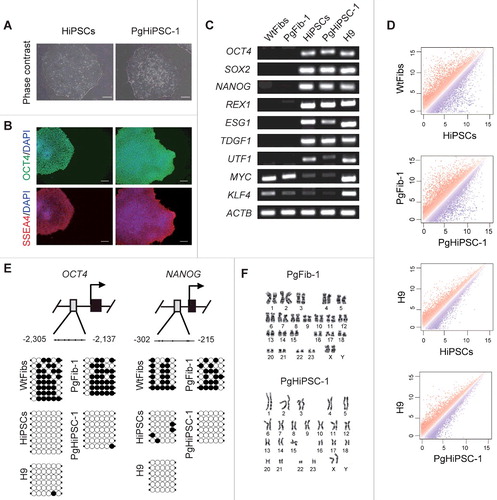
Figure 2. Analysis of expression and DNA methylation of known imprinted genes. (A) Relative mRNA expression levels of known paternally expressed genes in WtFibs, HiPSCs, PgFib-1, PgHiPSC-1, and H9. The data shown are averages of gene expression. (B) DNA methylation status of known paternally and maternally imprinted genes (H19, GRB10, and MEST) in PgFib-1, PgHiPSC-1, WtFibs, HiPSCs, and H9 was analyzed by bisulfite sequencing. Each line represents a separate clone. Black and white circles represent methylated and unmethylated CpGs, respectively.
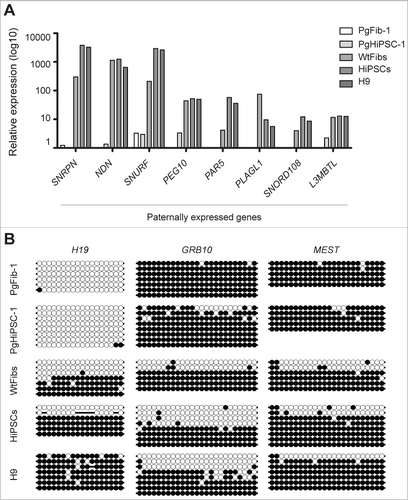
Table 1. A survey for candidate imprinted CpG sites