Figures & data
Figure 1. KDM4B abundance is significantly increased in periodontal diseased versus healthy tissues. Live Aa was injected subcutaneously into 12-week old C57BL/6 mice at the mid-sagittal region of the calvarium every day for 5d. Paraffin embedded sections were stained for F4/80, TRAP and KDM4B using immunohistochemistry, all of which were significantly upregulated in diseased versus healthy calvariae. 10x Images presented are representative of the data set. (a) In clinical periodontal specimens, the region of interest was defined as the connective tissue underlying the oral epithelium. Paraffin embedded sections were stained for KDM4E and KDM4B using immunohistochemistry, both of which were upregulated in diseased versus healthy patient tissues. 20x images are representative of the data set. (b) positive pixels quantified using color thresholding in imagej. Data are presented as mean ± SD. Significance was determined using a one-tailed Wilcoxon ranked sum test. Epithelium (E) Calvarial Bone (C) Brain (B) *P < 0.05, ***P < 0.001. Scale bars, 100 μm.
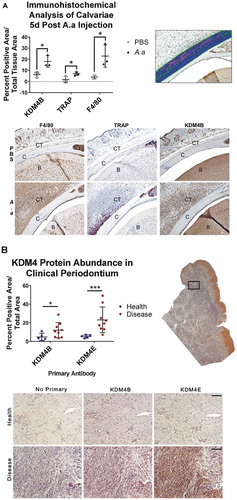
Figure 2. ML324 demonstrates inhibitory activity towards KDM4B and causes dose dependent immunosuppression. The KDM4A-E protein crystal structures were subjected to unbiased docking of ML324 where the top 30 conformers in the active site were used for analysis (a). KDM4A and C had no poses of ML324 dock into the active site of these enzymes, therefore this data is not displayed. Inhibition of KDM4B was assessed using an 11-point IC50 determination using the histone demethylase AlphaScreen (PerkinElmer) assay in triplicate resulting in an IC50 of 4.9 μM (b). The EC50 for immunosuppression using ML324 was determined to be 31 μM by measuring supernatant IL-6 protein following a 24h Aa-LPS stimulation with variable concentrations of ML324 in primary BMDM cells (c). Cells were treated for 1h with each indicated concentration of ML324, followed by Aa-LPS challenge for 24h. The data were normalized as a percentage of the maximal IL-6 response in response to LPS. Data for all panels are represented as mean ± SD. n = 4.
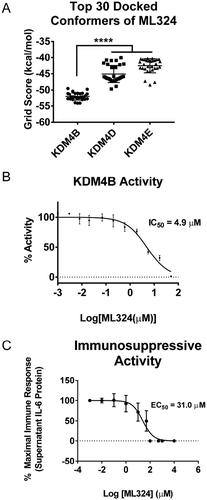
Figure 3. KDM4B inhibition significantly reduces the Aa-LPS-induced immune response in primary macrophages. Male and female murine bone marrow derived macrophages were pre-treated for 1h with the selective KDM4B inhibitor, ML324, a family-wide KDM4 inhibitor, JIB-04, and a KDM1A inhibitor, GSK-LSD1 or DMSO vehicle. Following drug treatment, cells were challenged with Aa-LPS for 8 and 24h, where gene expression and supernatant protein concentration were measured via rt-qPCR relative to GAPDH and ELISA relative to a standard curve. Data was normalized as a percentage of the maximal response (red) in each group for display. Refer to figure S1 for raw data and statistical significance. n = 4 per experiment, data is representative of 3 experiments.
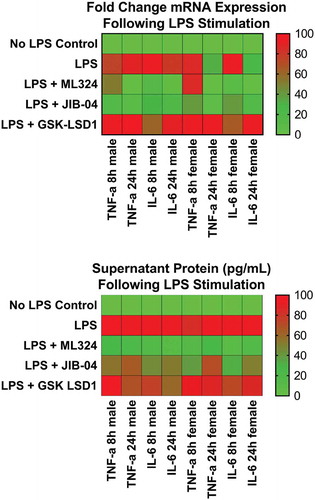
Figure 4. KDM4B inhibition via ML324 prevents osteoclast formation induced by either Aa-LPS or RANK-L. Murine bone marrow was differentiated into osteoclasts by supplementation of the hematopoietic compartment with M-CSF for 3 days and RANK-L + M-CSF for 2 days, where cells were rinsed and pre-treated for 1h with M-CSF and ML324 or DMSO vehicle followed by supplementation with RANK-L or Aa-LPS. After 72 hours, cells were fixed and stained for tartrate resistant acid phosphatase. 3 representative images were taken of each well, and TRAP+, multinucleated cells were counted in each field. 10x representative images of M1 and F2 are displayed (a). Each mouse (M1, M2 = male; F1, F2 = female) independently showed a significant increase in osteoclast formation in response to LPS or RANK-L alone compared to PBS controls, but no significant difference in osteoclast formation was observed between PBS and ML324 + LPS or ML324 + RANK-L treated cells (b). Data is presented as the mean number of osteoclasts in each field ± SD. Statistical significance was determined using a paired Friedman test with multiple comparisons at an α = 0.05. *P < 0.03 n = 3 fields per well, 2 wells per group, per mouse.
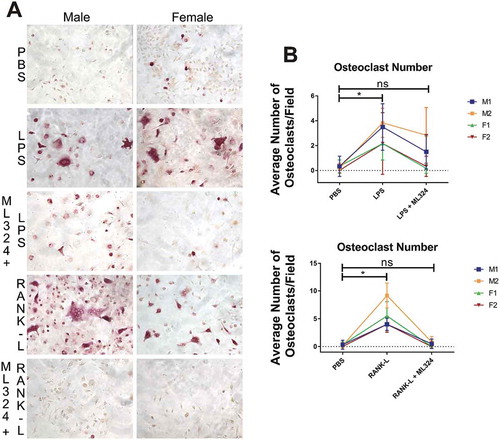
Figure 5. KDM4B inhibition mediated immunosuppressive effect is indirect, requiring new protein synthesis and increased KDM1A activity. New protein synthesis is required for the immunosuppressive effects of ML324 on the LPS-induced immune response (a). Cells were pre-treated with cycloheximide (5ug/mL) for 4 h with or without ML324 (50 μM) followed by Aa-LPS challenge (100 ng/mL) for 16h where RNA was collected and analyzed using qRT-PCR to measure IL-6 expression compared to GAPDH as an endogenous control. Statistical significance was determined using a repeated measures ANOVA with multiple comparisons. Demethylation at H3K4 is significantly decreased in cells following LPS treatment, but the effect is reversed upon addition of ML324. (b) RAW264.7 cells were pre-treated for 1h with ML324 followed by Aa-LPS challenge for 24h where H3K4 mono-methylation was measured in fixed cells using immunofluorescent antibody to H3K4me at 10x. KDM1A activity is significantly decreased following Aa-LPS treatment alone, but in combination with ML324 pretreatment, KDM1A activity is increased. n = 3 wells, 48 fields/well, **P < 0.01. Data are represented as mean ± SD. Statistical significance was determined using one-way ANOVA with multiple comparisons where the outcome was log transformed.
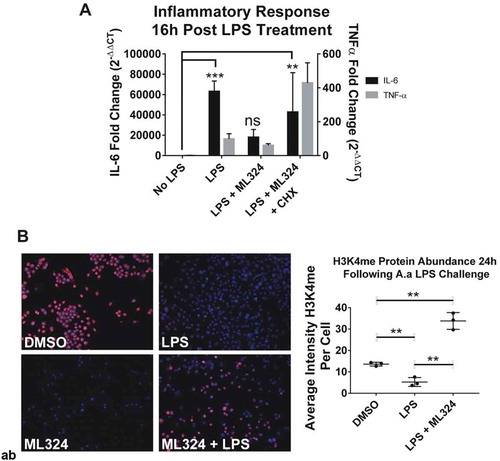
Figure 6. A schematic illustration of the mechanism of KDM4B inhibition-mediated immunosuppression. KDM4B activity inactivates KDM1A and allows for active transcription of pro-inflammatory cytokines while KDM4B inhibition allows for active demethylation by KDM1A, reducing pro-inflammatory cytokine production.