Figures & data
Figure 1. Histone lysine PTMs and oncogenic mutations. (a) Schematic representation of writers, readers and erasers targeting lysine residues in histone tails of the nucleosome. (b) Residues of the H3 tail and mutations that have been linked to cancer are colored green and red, respectively.
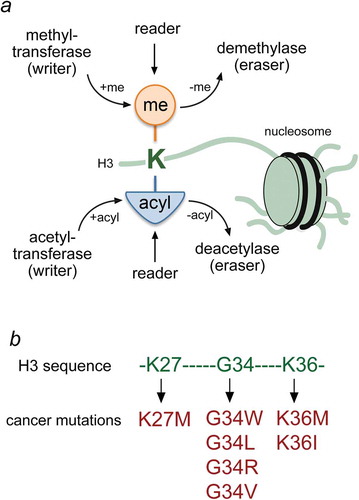
Figure 2. Structural basis for entrapping H3K36M in the SETD2 active site. (a) The crystal structure of the SETD2 catalytic domain (SET in white and Post-SET in pink) in complex with the H3K36M peptide (orange) and SAH (blue). SETD2 CD is shown in a surface model with the K36M-binding pocket residues and the G34-binding site residues colored yellow and light green, respectively. (b) A close view of the ribbon diagram of the H3K36M-CD-SAH complex structure. PDB ID code: 5JJY [Citation70].
![Figure 2. Structural basis for entrapping H3K36M in the SETD2 active site. (a) The crystal structure of the SETD2 catalytic domain (SET in white and Post-SET in pink) in complex with the H3K36M peptide (orange) and SAH (blue). SETD2 CD is shown in a surface model with the K36M-binding pocket residues and the G34-binding site residues colored yellow and light green, respectively. (b) A close view of the ribbon diagram of the H3K36M-CD-SAH complex structure. PDB ID code: 5JJY [Citation70].](/cms/asset/fb06afb5-d6e5-461d-97b1-cf94b9d10658/kepi_a_1503491_f0002_oc.jpg)
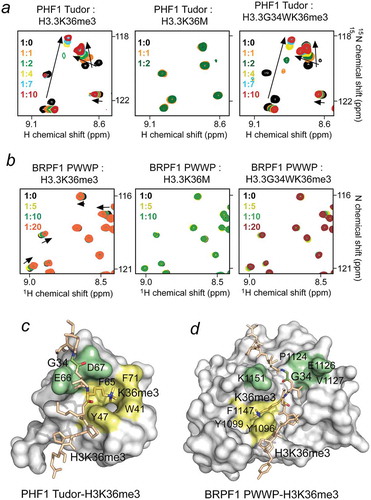
Figure 3. Oncohistone mutations impact binding of the H3K36me3 readers. (a, b) Superimposed 1H,15N HSQC spectra of PHF1 Tudor (a) and BRPF1 PWWP (b) recorded while H3H36me3, H3K36M and H3G34WK36me3 (residues 28–40 of H3.3) peptides were added stepwise. The spectra are color coded according to protein:peptide molar ratio. The structures of the H3K36me3-bound PHF1 Tudor domain and H3K36me3-bound BRPF1 PWWP domain are shown as surface models with the K36M-binding pocket residues and the G34-binding site residues colored yellow and light green, respectively. PDB ID codes: 4HCZ and 2X4X.