Figures & data
Figure 1. DNA methylation levels of 12 genes in the in vitro model. HFK, human foreskin keratinocytes (n = 3); Early, early immortal passages of FK16A, FK16B, FK18A and FK18B p32–p52 (n = 4); Late, late immortal passages of FK16A, FK16B, FK18A and FK18B p129-p156 (n = 4); CCL, cancer cell lines (n = 3). Each of the individual cell lines is indicated by a different symbol. Only for the FK16A, FK16B, FK18A, and FK18B in the 2nd and 3rd column the symbols refer to the early and late passages of the same cell line.
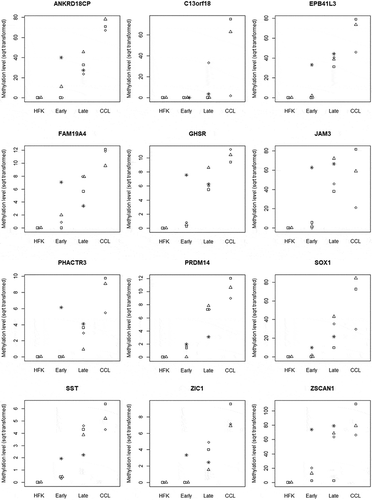
Figure 2. DNA methylation level distributions in hrHPV-positive cervical scrapings across histological subgroups for 12 genes. Normal, hrHPV-positive controls (n = 26); CIN2 and CIN3, cervical intraepithelial neoplasia grade 2 (n = 20) and 3 (n = 53); (mi)Ca, micro-invasive cervical cancer (n = 14). *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
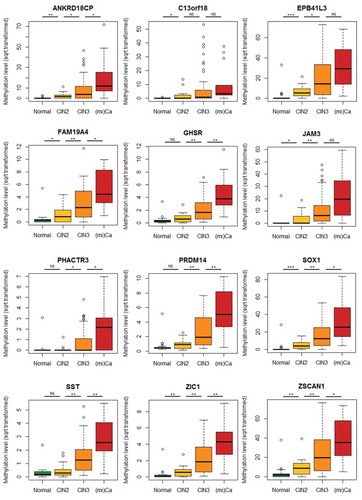
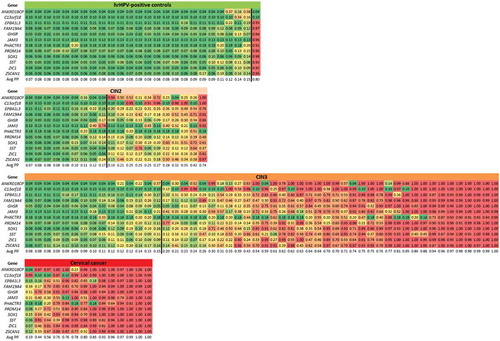
Figure 3. DNA methylation signature of 12 genes in hrHPV-positive cervical scrapings. Predicted probabilities are shown for 12 genes in the different histological subgroups and colored from green (predicted probability of 0) to red (predicted probability of 1). In each group, samples are ordered based on their average predicted probability (Avg PP). The black line indicates the cut-off for a cancer-like methylation-high pattern at ≥0.19.