Figures & data
Figure 1. Overview of the chromosomal region 14q32.
The figure gives an overview of the imprinted gene cluster on chromosome 14q32. Maternally expressed genes are indicated in red, paternally expressed genes in blue. The region harbors three differentially methylated regions (DMRs): the MEG3/DLK1:IG-DMR and the MEG3:TSS-DMR that are methylated on the paternal allele (pat) and the MEG8:Int2-DMR which is methylated on the maternal allele (mat). The methylation status is indicated by filled (methylated) and empty (unmethylated) lollipops. Expression of the genes is indicated by arrows.
The situation in TS14 patients is given below. The MEG3/DLK1:IG-DMR and the MEG3:TSS-DMR are unmethylated on both alleles while the MEG8:Int2-DMR is methylated on both alleles. This leads to an increased expression of the maternally expressed genes (indicated by bold gene names) while the paternally expressed genes are silenced (indicated by grey gene names).
Modified from Beygo et al. 2017 [Citation8].
![Figure 1. Overview of the chromosomal region 14q32.The figure gives an overview of the imprinted gene cluster on chromosome 14q32. Maternally expressed genes are indicated in red, paternally expressed genes in blue. The region harbors three differentially methylated regions (DMRs): the MEG3/DLK1:IG-DMR and the MEG3:TSS-DMR that are methylated on the paternal allele (pat) and the MEG8:Int2-DMR which is methylated on the maternal allele (mat). The methylation status is indicated by filled (methylated) and empty (unmethylated) lollipops. Expression of the genes is indicated by arrows.The situation in TS14 patients is given below. The MEG3/DLK1:IG-DMR and the MEG3:TSS-DMR are unmethylated on both alleles while the MEG8:Int2-DMR is methylated on both alleles. This leads to an increased expression of the maternally expressed genes (indicated by bold gene names) while the paternally expressed genes are silenced (indicated by grey gene names).Modified from Beygo et al. 2017 [Citation8].](/cms/asset/41dbab83-2e33-4616-a7b4-1f32e7344c39/kepi_a_1514233_f0001_c.jpg)
Figure 2. Genomic imprinting and the occurrence of imprint errors.
This scheme shows the so called imprint life cycle exemplarily for the germline-derived MEG3/DLK1:IG-DMR on chromosome 14q32. The normal process of genomic imprinting is depicted in the column on the left-hand side. The methylation imprints of the DMRs are indicated by lollipops (filled – methylated status of the DMR, empty – unmethylated status of the DMR) and shown for the maternal (red) and paternal (blue) alleles.
Somatic cells with a normal methylation imprint are shown at the top. These imprints are then erased in the primordial germ cells. The asterisk on the maternal allele indicates an epigenetic mark that prevents DNA methylation on the normally unmethylated maternal allele e.g., H3K4me2 or H3K4me3. The methylation imprints are then newly established in the male and female germ cells, respectively and are subsequently maintained in the zygote and through all following cell divisions.
The other three columns display errors in imprint erasure, imprint establishment and imprint maintenance and their outcome in regard to chromosome 14.
A failure to erase the maternal imprint in the paternal primordial germ cells would lead to a maternal epigenotype on the paternal allele. In these cases, a TS14 ID patient would have inherited the allele harboring the ID exclusively from the paternal grandmother. (Other marks may apply for mature sperm because the genome is largely devoid of histones.)
In case the imprint erasure (second column) works correct but the imprint is not established in the male germ cells. Here, the ID allele could be inherited either from the paternal grandmother or the grandfather. The same holds true for errors in imprint maintenance. Additionally, mosaic IDs occur when the failure took place after fertilization in the early embryo so that only a subset of cells harbor an ID while the other cells have a normal epigenotype.
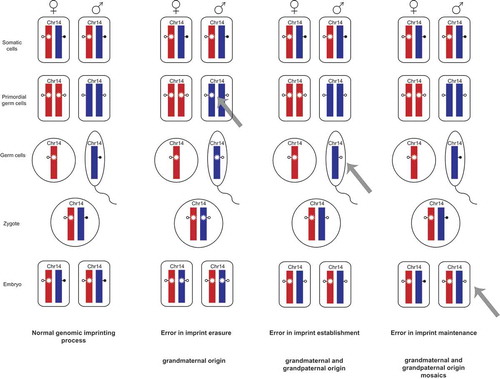
Table 1. Summary of the results.
Figure 3. Methylation analysis and model of imprint maintenance error for a mosaic imprinting defect.
Methylation results (upper line) obtained by deep bisulfite sequencing for the three DMRs on 14q32 are depicted for patient 12 who has a mosaic imprinting defect. The paternally methylated MEG3/DLK1:IG-DMR and MEG3:TSS-DMR show a hypomethylation with methylation levels of 38.2% (CpGs6-8 28.7%) and 23.5%, respectively. In accordance with these results is the hypermethylation of the maternally methylated MEG8:Int2-DMR, which exhibits a methylation level of 79.6%.
The scheme below shows the theoretical emergence of a mosaic imprinting defect due to a postzygotic error in imprint maintenance. At first, the methylation imprint is lost at the MEG3/DLK1:IG-DMR. As this DMR governs the MEG3:TSS-DMR in a hierarchical fashion it can be assumed that the methylation is lost at the MEG3:TSS-DMR subsequently. By this, transcription of a hypothesized long transcript starting at the MEG3 promoter and running through the MEG8:Int2-DMR can occur from the paternal allele [Citation8]. This transcription leads to the establishment of methylation at the paternal MEG8:Int2-DMR and its hypermethylation.
![Figure 3. Methylation analysis and model of imprint maintenance error for a mosaic imprinting defect.Methylation results (upper line) obtained by deep bisulfite sequencing for the three DMRs on 14q32 are depicted for patient 12 who has a mosaic imprinting defect. The paternally methylated MEG3/DLK1:IG-DMR and MEG3:TSS-DMR show a hypomethylation with methylation levels of 38.2% (CpGs6-8 28.7%) and 23.5%, respectively. In accordance with these results is the hypermethylation of the maternally methylated MEG8:Int2-DMR, which exhibits a methylation level of 79.6%.The scheme below shows the theoretical emergence of a mosaic imprinting defect due to a postzygotic error in imprint maintenance. At first, the methylation imprint is lost at the MEG3/DLK1:IG-DMR. As this DMR governs the MEG3:TSS-DMR in a hierarchical fashion it can be assumed that the methylation is lost at the MEG3:TSS-DMR subsequently. By this, transcription of a hypothesized long transcript starting at the MEG3 promoter and running through the MEG8:Int2-DMR can occur from the paternal allele [Citation8]. This transcription leads to the establishment of methylation at the paternal MEG8:Int2-DMR and its hypermethylation.](/cms/asset/90fe049d-db94-47fa-829c-e96258939d08/kepi_a_1514233_f0003_c.jpg)
