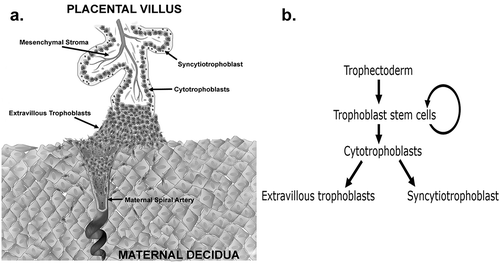
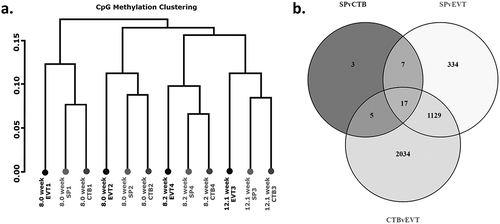
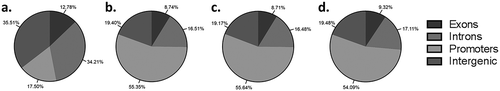
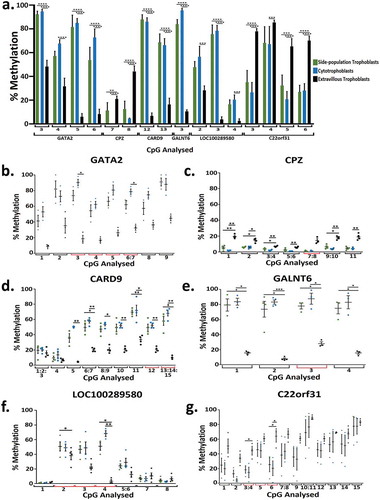
Figures & data
Table 1. Table of sequenom massARRAY primers.
Table 2. Table of microRNA primers.
Table 3. Summary table of differential methylation between trophoblast populations.
Table 4. Table of differentially methylated genes regulating a stem cell phenotype and/or cell cycle regulation and proliferation in side-population compared to cytotrophoblasts or extravillous trophoblasts.
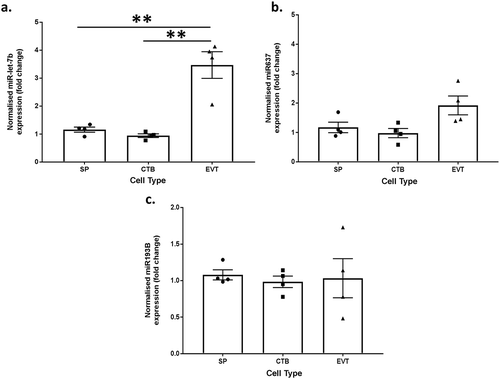
Table 5. Table of miRNA involved in signalling pathways regulating stem cells.
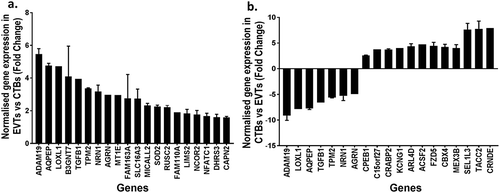
Table 6. Table of differentially methylated genes related to EMT, and/or metastatic cancers were up-regulated in extravillous trophoblasts compared to cytotrophoblasts.