Figures & data
Table 1. Summary of differentially methylated CpG sites in whole blood and different cell types.
Table 2. pSLE-specific DNAm signature.
Figure 1. PCA of the DNA methylation dataset in all 145 samples.
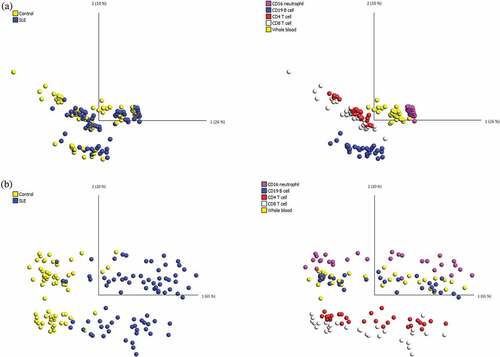
Figure 2. Venn diagram showing differentially methylated CpG sites across different cell lineages.
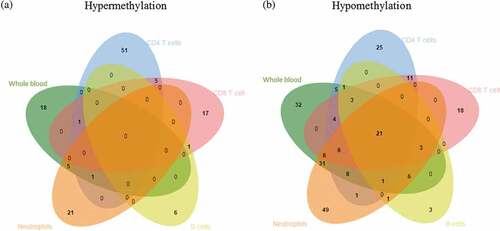
Figure 3. Heatmap of the pSLE-specific DNAm signature.
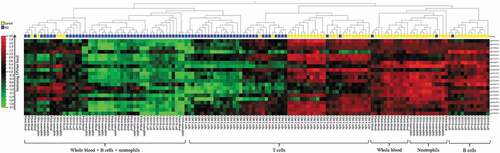
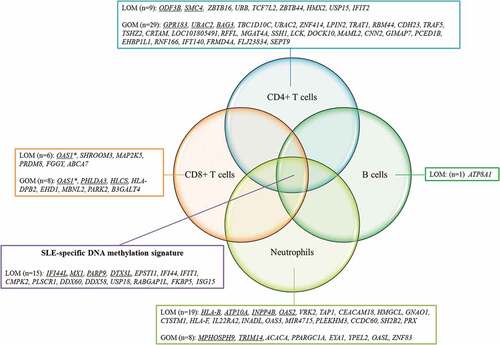
Figure 4. Lineage-specific DNAm changes in pSLE patients.