Figures & data
Figure 1. Reactivation of methylated P16 alleles in H1299 cells 6 days after P16-TET transient transfection. (a) Core sequences of the catalytic domain (CD) of the wildtype TET1 gene and its T > C H1671Y-mutant counterpart. The P16-TET pcDNA3.1 and pTRIPZ ‘Tet-on’ expression vectors are also illustrated and used in transient and stable transfection studies, respectively. (b) The results of qRT-PCR to detect the P16 mRNA levels in H1299 cells transiently transfected with the pcDNA3.1 empty control, P16-TET and its H1671Y mutant counterpart vectors. (c) Immunofluorescence staining with the mouse monoclonal antibody against the human P16 protein (Ventana Roche-E6H4, USA). These experiments were performed in triplicate and repeated at least one time. Bar: 50 μm.
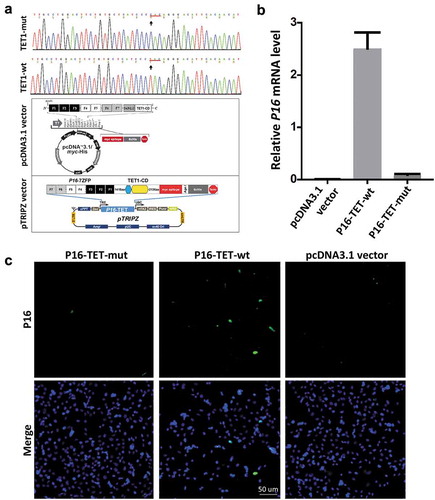
Figure 2. P16-TET induces hydroxymethylation of P16 CpG islands and reactivates expression of methylated P16 alleles in H1299 cells. (a) TAB-MSP analysis for detecting hydroxymethylated (H)- and nonhydroxymethylated (N)-P16 CpG alleles in H1299 cells stably transfected with P16-TET or empty control vector after doxycycline treatment. The MSP analysis results were also listed. Genomic DNA from RKO and BGC823 cells was used as P16M and P16U controls in the MSP assays, respectively. (b) Western blot analysis for detecting the P16 protein; Dox (±): with or without the doxycycline treatment (final conc. 0.25 μg/mL). Proteins from BGC823 cells were used as a P16U/active control. (c) qRT-PCR results for detecting P16 mRNA levels relative to Alu RNA levels; (d) Immunofluorescence confocal analysis for detecting P16 expression. P16-positive cells with P16-TET (Myc-tag) expression are highlighted with white dash cycles; P16-negative cells with P16-TET (Myc-tag) expression are highlighted with pink dash cycles. Bar: 30 μm.
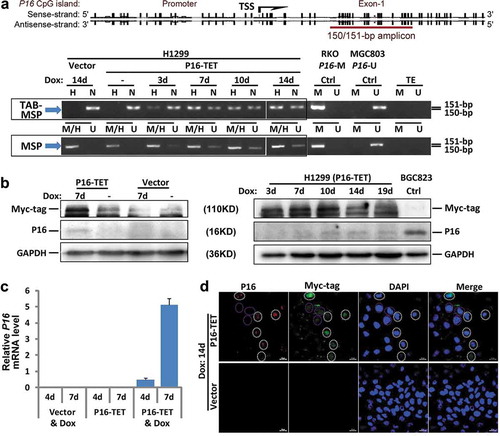
Figure 3. P16-TET induces hydroxymethylation of P16 CpG islands and reactivates expression of methylated-P16 alleles in AGS cells. (a) Bisulphite-DHPLC analysis for detecting methylated-P16 (P16M) and unmethylated-P16 (P16U) PCR products for the exon-1 antisense strand in P16-TET-transfected AGS cells with different doxycycline induction times. (b) The TAB-DHPLC analysis detected the hydroxymethylated P16 (P16H) PCR products and nonhydroxymethylated P16 (P16N) PCR products. (c and d) Bisulphite and TAB sequencing for detecting 5mC and 5hmC sites, respectively, in the same PCR products as were analysed by DHPLC. (e and f) The results of RT-PCR and Western blot analysis for detecting P16 reactivation in AGS cells.
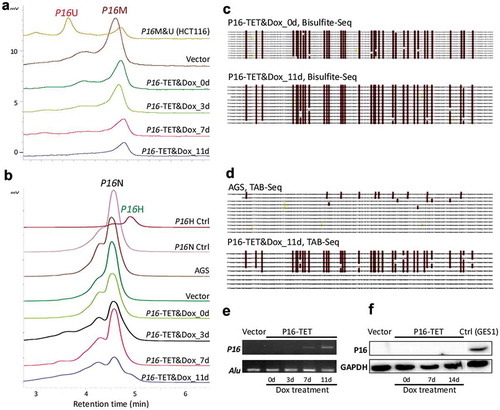
Figure 4. Characterization of P16H in FACS-sorted subpopulations of H1299 cells with various levels of of P16 expression reactivation. (a) FACS sorting of P16-TET stably transfected H1299 cells with and without Dox treatment. The confocal images of the P16 protein staining status are also attached. Bar: 30 μm. (b) Detection of the DNA hydroxymethylation status of P16 alleles in various FACS-sorted H1299 subpopulations with strong, weak, and no P16 immunostaining (P16(+)/(±)/(-)) in the TAB-MSP analysis. (c) The results of TAB sequencing for the P16 CpG islands in the P16-negative subpopulation.
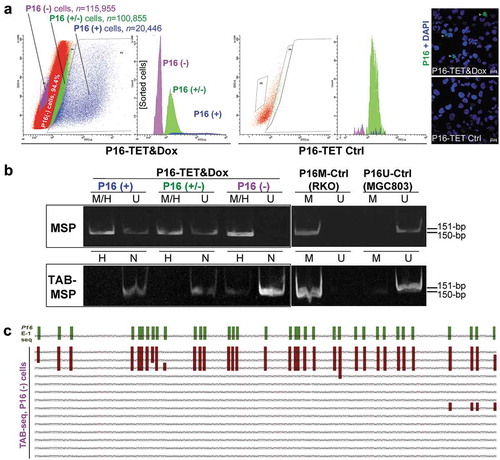
Figure 5. Characterization of P16H in FACS-sorted subpopulations of AGS cells with various levels of P16 expression reactivation. (a) FACS-sorting of P16-TET stably transfected AGS cells with and without DAC treatment. The confocal images of the P16 protein staining status are also attached. Bar: 50 μm. The sorted AGS cells not treated with doxycycline only expressed P16-TET at the baseline level. (b) Detection of the DNA hydroxymethylation status of P16 alleles in various FACS sorted AGS subpopulations with strong, weak, and no P16-immunostaining (P16(+)/(±)/(-)) in the TAB-MSP analysis. (c) The results of TAB-DHPLC for the P16 CpG islands in three subpopulations. (d) The results of TAB sequencing for the antisense strand of P16 exon-1 in P16(+) and P16(-) subpopulations.
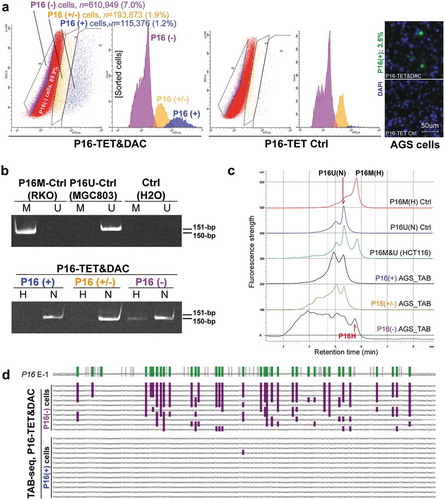
Figure 6. Effects of knockdown of TET1/2/3 on reactivation of the P16 gene by DNA methylation inhibitor DAC and P16-specific artificial transcription factor (P16-ATF) in HCT116 cells. After treated with the TET-specific siRNAs for 48 hrs, these cells were transiently transfected with the P16-ATF vector for 24 hrs, and then treated with DAC (final conc. 20 nmol/L) for 48 hrs. (a) The relative mRNA levels of TET1/2/3 in gene-specific siRNA treated cells in qRT-PCR ananlysis; (b) The confocal images of the P16 protein staining status for various group cells.
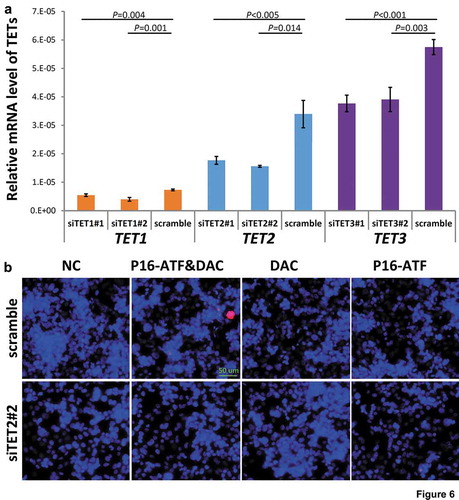
Figure 7. Effects of P16H on the proliferation of H1299 cells in vitro and in vivo. (a) Cell proliferation curves for H1299 cells with and without P16-TET expression in a live content kinetic imaging platform; (b) Comparison of weights of H1299 tumour xenografts with and without stable P16-TET transfection in SCID mice; (c and d) Images of xenografts on the 50th experimental day.
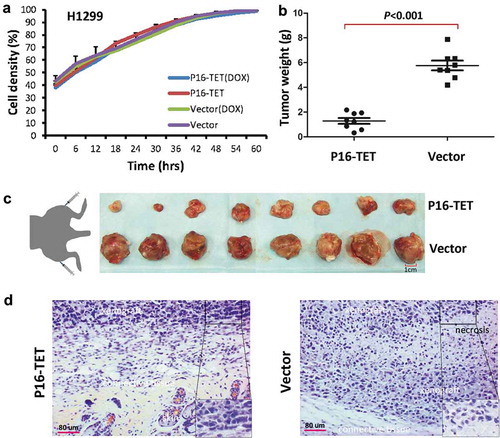
Table 1. Sequences of oligonucleotides used as primers in various PCR-based assays.
