Figures & data
Table 1. Characteristics of the study population.
Table 2. Primers used for RT-qPCR gene expression analysis.
Figure 1. Quantification of global DNA methylation and hydroxymethylation. DNA methylation levels in LINE-1 repeat elements, as a measure of global 5-mC, and global DNA hydroxymethylation (5-hmC) levels were determined in vapers, smokers, and controls by ELISA, as described in the text. All samples were assayed in triplicate (for 5-mC) and in duplicate (for 5-hmC). Distribution of data within each group is shown by a combination of scatter plots (to display individual values) and box and whisker plots (to highlight the minimum, first quartile, median, third quartile, and maximum values as well as the means and outlier(s) (if any)). In the scatter plots, identical values are overlaid and presented as a single circle (‘°’). In the box and whisker plots, the ‘lower’ and ‘upper’ edges of boxes represent the 1st and 3rd quartiles, respectively (25 and 75 percentiles, resp.). Horizontal lines within the boxes represent the medians (2nd quartile or 50 percentile) and small crosses (‘x’) indicate the mean values. The ‘lower’ and ‘upper’ vertical lines extending from the boxes, also known as the ‘whiskers’, represent the lowest and highest data points, respectively, excluding any outliers (minimum and maximum values, resp.). Comparison between groups was done by the Wilcoxon Rank-Sum test. Panels A and B show the quantification results for 5-mC and 5-hmC, respectively.
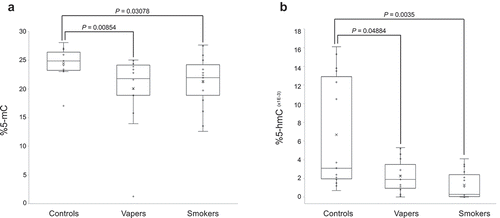
Figure 2. Expression analysis of DNMTs and TETs. Expression level of DNMTs (DNMT1, DNMT3A, and DNMT3B) and TETs (TET1, TET2, and TET3), catalysing DNA methylation and hydroxymethylation, respectively, were measured in vapers, smokers, and controls by RT-qPCR, as described in the text. All samples were assayed in triplicate. Distribution of data within each group is shown by a combination of scatter plots and box and whisker plots, as described in the legend for . Panels A and B show the results of expression analysis for DNMTs and TETs, respectively. The modest changes in the expression level of DNMTs or TETs observed in vapers and smokers relative to controls or relative to each other did not reach a statistically significant level (P < 0.05), as determined by the Wilcoxon Rank-Sum test.
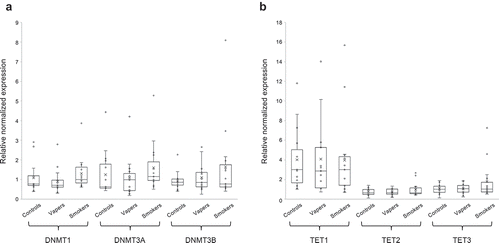
Figure 3. Measurements of plasma cotinine, exhaled CO and COHb. Concentrations of plasma cotinine were measured by ELISA, and the exhaled CO and COHb levels were determined by a breath CO monitor, as described in the text. Distribution of data within each group is shown by a combination of scatter plots and box and whisker plots, as described in the legend for . Comparison between groups was done by the Wilcoxon Rank-Sum test. Panel A shows the results of cotinine measurement, whereas panels B and C display the quantification data for CO and COHb, respectively.
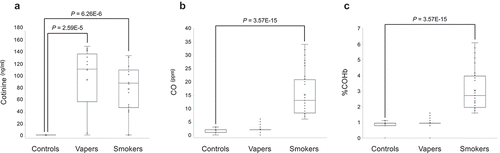
Table 3. Relationship of vaping/smoking indices and effect variables.
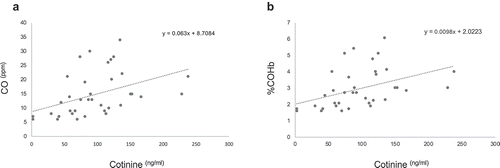
Figure 4. Correlation analysis of plasma cotinine and breath CO and COHb. The concentrations of plasma cotinine in smokers were directly and statistically significantly correlated to their exhaled breath CO (panel A) and %COHb (panel B) (r = 039,154, P = 9.78E-6 and r = 039,153, P = 9.78E-6, respectively; Spearman Rank correlation test).