Figures & data
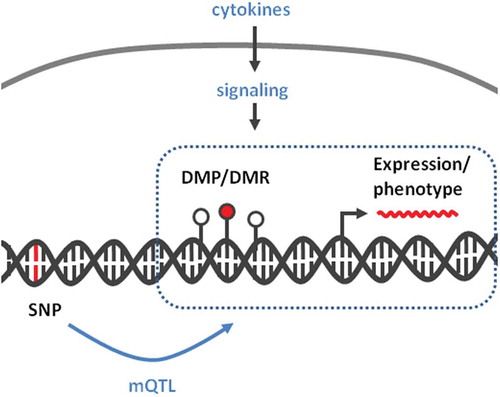
Table 1. Dataset characteristics.
Table 2. Samples used in the study.
Table 3. Top DMPs.
Figure 1. DNA methylation distinguishes IBD from healthy intestinal epithelial cells.
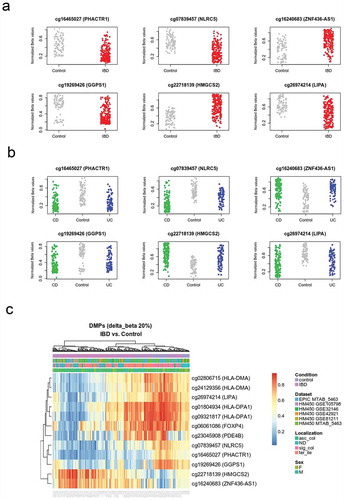
Table 4. Top DMRs.
Figure 2. Mean DNA methylation and variability distinguishes IBD from healthy intestinal epithelial cells.
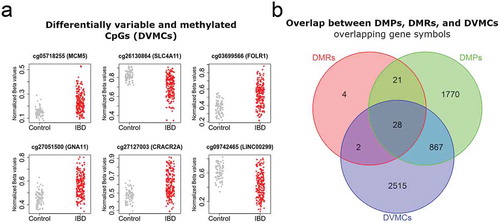
Table 5. Pathway analysis.
Figure 3. Genomic distribution of IBD-related DMPs.
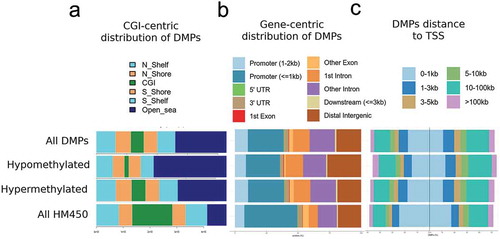
Figure 4. Genomic distances between IBD-related DMPs and known risk SNPs.
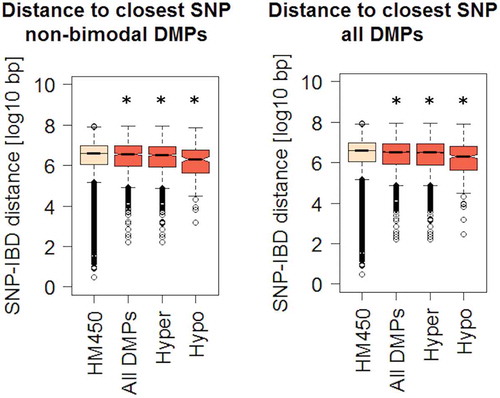
Table 6. IBD DMPs previously identified to be differentially methylated in both CeD duodenal epithelia and immune fractions.
