Figures & data
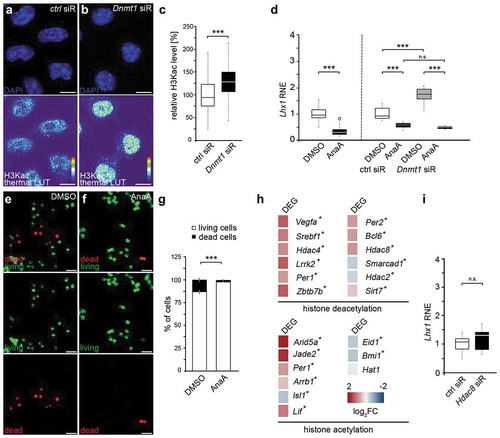
Figure 1. Lhx1 transcription is controlled by DNMT1, but not through its DNA methylation activity
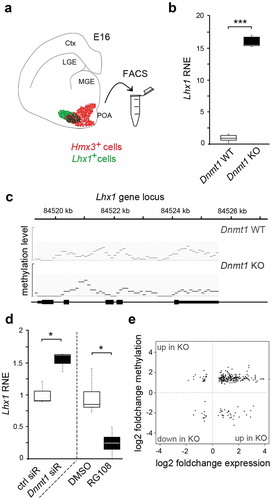
Figure 2. DNMT1-dependent modulation of repressive and permissive histone lysine trimethylation contributes to Lhx1 expression control
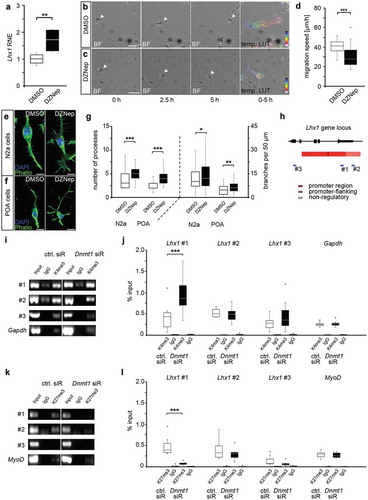
Figure 3. Lhx1 transcription is modulated by DNMT1-dependent histone acetylation and deacetylation processes