Figures & data
Figure 1. Age-related expression of miRNAs indicated in silico as interacting with SIRT6 mRNA in PBMCs. (a) Levels of miR-125b-5p are not affected by age. (b) Levels of miR-34a-5p, miR-103-3p, miR-125a-5p, miR-181a-2-3p, miR-186-5p, miR-342-5p, miR-776-3p and Let-7c are significantly affected by age. Results are presented as median with interquartile range. Statistical analysis was performed with the Kruskal-Wallis test and Dunn’s multiple comparisons test. Y: young (mean age 27.5 ± 4.4 years), E: middle-aged (65.4 ± 3.3 years), LL: long-lived (93.9 ± 3.6 years)
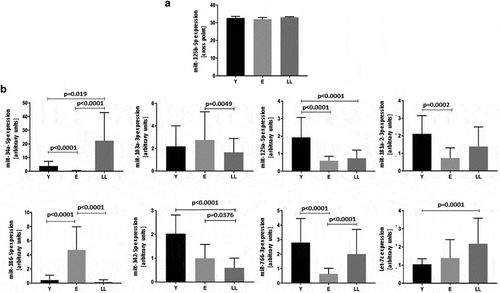
Figure 2. Effects of interaction of the 3´UTR of SIRT6 mRNA with miR-186-5p. (a) Co-transfection of the pmirGLO_SIRT6 vector containing the full-length 3´UTR of SIRT6 mRNA and the firefly luciferase reporter gene with pre-miR-186-5p results in a significant decrease in luminescence as compared to co-transfection with the pre-neg-miR negative control (neg1). Results are presented as mean with standard deviation. Statistical analysis was performed with a two-tailed Student t-test. (b) Transfection of HEK293 cells with pre-miR-186-5p results in a decrease in the expression of endogenous SIRT6 protein. Hsp90 – heat shock protein 90, control protein expression of which is not affected by miR-186-5p. The Western blot analysis was performed once, and the results of densitometric scanning are provided in Supplementary Table S2
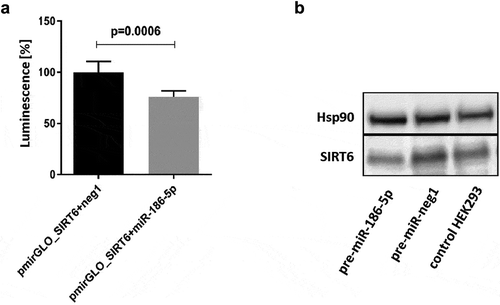
Table 1. Correlation of miRNAs expression with the expression of SIRT6 in human PBMCs
Figure 3. The interaction network between proteins, each of which is involved in the regulation of immunity, apoptosis, and ageing (bold circles), which are also direct or indirect SIRT6 interactors. The network was constructed using the STRING functional protein association network version 10.5 (https://string-db.org/) with the minimum required interaction score of 0.4 (medium confidence). To fill ‘gaps’ between SIRT6 and these proteins, the program was allowed to include up to 5 additional interactors in the first shell
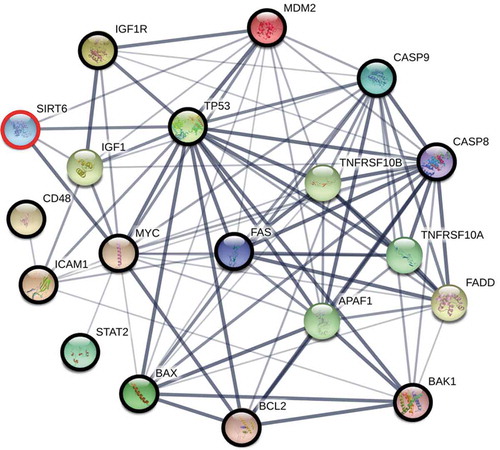
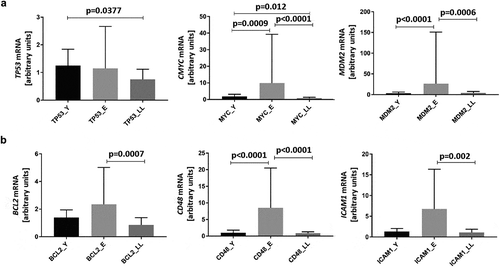
Figure 4. Age-related mRNA expression of proteins interacting with SIRT6 in PBMCs. Results, normalized against the expression of the β-actin gene (ACTB), are presented in arbitrary units as median mRNA levels with interquartile range. Statistical analysis was performed with the Kruskal-Wallis test and Dunn’s multiple comparisons test. (a) Direct SIRT6 interactors TP53, MYC, and MDM2. (b) Indirect SIRT6 interactors BCL2, CD48, and ICAM1. Y: young (mean age 27.5 ± 4.4 years), E: middle-aged (65.4 ± 3.3 years), LL: long-lived (93.9 ± 3.6 years)