Figures & data
Figure 1. Potential redox of glutathione and vitamin C concentrations depending of nutritive modalities
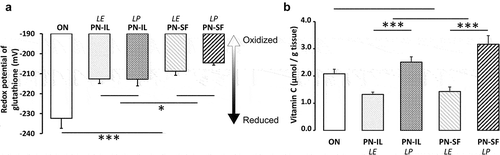
Table 1. Hepatic GSH and GSSG levels depending on nutritive modalities
Table 2. Characteristics of lipid emulsions
Figure 2. DNA methylation depending of nutritive modalities
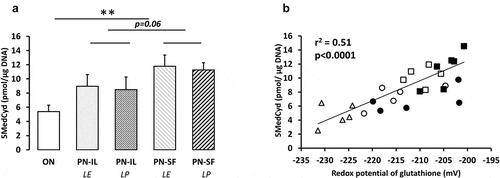
Figure 3. DNA methylation activity, levels of SAM and SAH depending of nutritive modalities
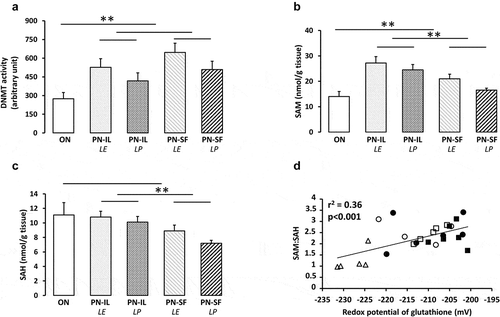
Figure 4. DNMT1 and DNMT3b protein levels depending of nutritive modalities
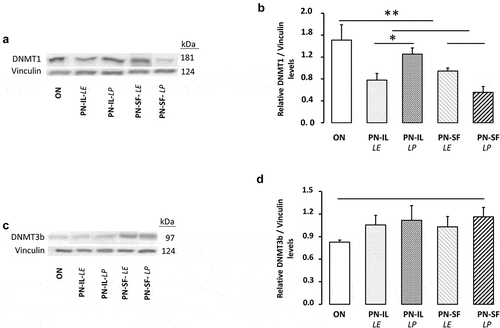
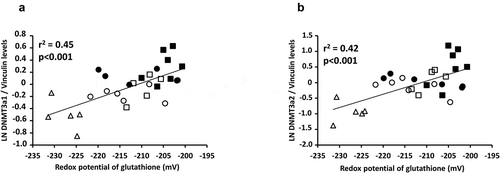
Figure 5. DNMT3a protein levels depending of nutritive modalities
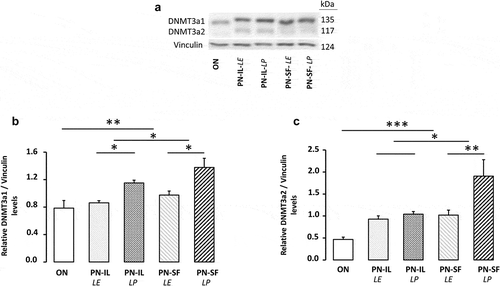
Figure 6. Relationships between DNMT3a and redox potential of glutathione