Figures & data
Figure 1. ATAC-seq identified peaks in IL-1-stimulated chondrocytes
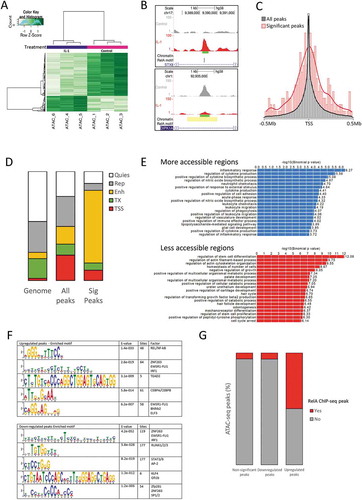
Figure 2. Correlation between differentially accessible chromatin regions and gene expression changes
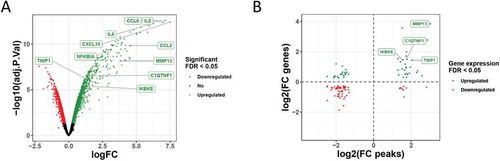
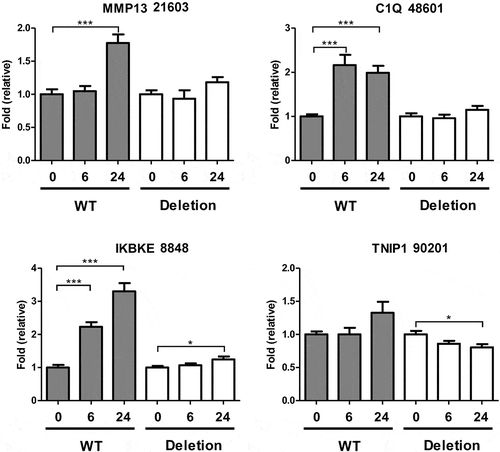
Figure 3. Effect of Cas9-mediated putative enhancer deletion on IL-1-induced gene expression
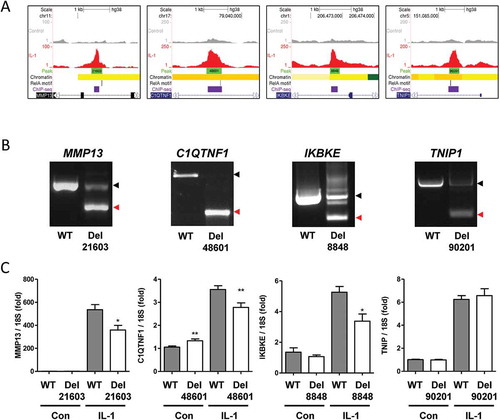
Figure 4. Effect of enhancer-targetted Cas9-VPR on gene expression
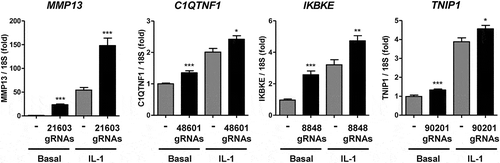
Figure 5. Enhancer activity of differentially accessible chromatin regions