Figures & data
Figure 1. Clinical course and mutations detected in I-2(a) and II-1(b). Germline mutation in DNMT3A P709S (black) and predominant somatic mutations in IDH2 R172K and ASXL1 P808fs (grey) that were detected within bone marrow samples at various timepoints (blue) along with the regimens and protocols given (green): CIA (induction of clofarabine, idarubicin and cytarabine), SGI-110 (DNMT inhibitor), BL-8040 (CXCR4 inhibitor), TH302 (Evofosfamide, replication inhibitor), AZD1208 (Pim-kinase inhibitor), Erlotinib (epidermal growth factor receptor (EGFR) inhibitor), PRI-724 (Wnt inhibitor), J0894 + J9100 (Decitabine + Cytarabine), AG221 x 2 cycles (IDH2 inhibitor), Trametinib (MEK inhibitor), APTO-253 (MTF-1 inhibitor) are shown
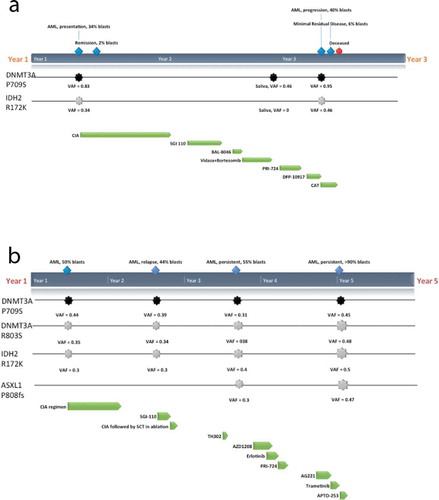
Figure 2. Position of the germline DNMT3A P709 within (a) the SAM-dependent methyltransferase (arrow) (cBioPortal) [Citation45,Citation46] and (b) the substrate-binding pocket of the DNA methyltransferase in the crystal structure (CN3D) [Citation47] and its positional relationship to the R882 canonical mutation
![Figure 2. Position of the germline DNMT3A P709 within (a) the SAM-dependent methyltransferase (arrow) (cBioPortal) [Citation45,Citation46] and (b) the substrate-binding pocket of the DNA methyltransferase in the crystal structure (CN3D) [Citation47] and its positional relationship to the R882 canonical mutation](/cms/asset/f3326a6b-f32c-41f0-ad03-3ac6483999c7/kepi_a_1809871_f0002_c.jpg)
Figure 3. The P705S mutation severely impairs the ability of Dnmt3a to methylate DNA
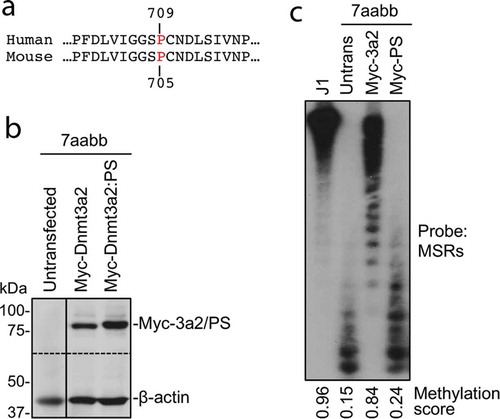
Figure 4. The Dnmt3a2:P705S mutant protein interacts with and antagonizes wild-type Dnmt3a2
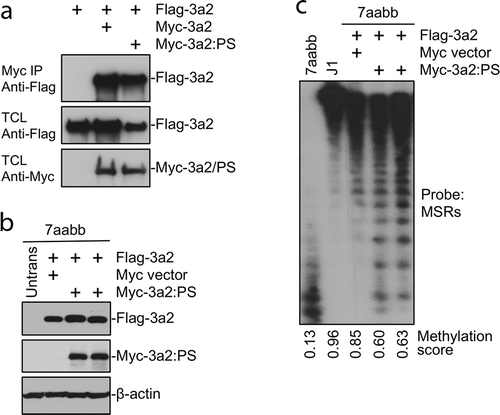
Figure 5. DNMT3A p.P709S is associated with decreased DNA methylation in germline samples