Figures & data
Table 1. Summary characteristics of retinoblastoma and peripheral blood DNA samples tested in this study CpGs counted as methylated includes any CpG with at least partial methylation (>10% per allele). (PB=peripheral blood). 1011 (homo c.1363C>T), 1012 (homo c.1814+1291T>G), 1013 (homo c.844G>T), 1014 (homo c.1215+1G>A), 1015 (c.2663+1G>A/c.-?1814+?del). *DNA methylation boundary location where CpGs are methylated up to and including boundary site with weak or no methylation downstream. Specimens PB=peripheral blood, T=tumour
Figure 1. Known promoter elements and CTCF binding motif with DNA contact regions in black. Numbers indicate CpG location relative to translation start site (arrowed). RB1 gene and LINC00441 transcription start sites also arrowed. RB1 5ʹ untranslated region in grey. Core promoter (CP) and ALU repeat sequences (MIR1 and H1PA8) highlighted in black.
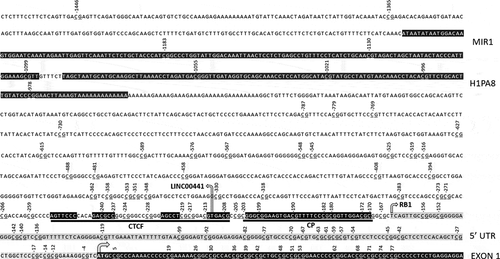
Figure 2. (a) Exons, 5ʹUTR, CTCF binding site, Core promoter region and long non-coding RNA exons. (b) CTCF binding site (boxed, contact regions in black), RB1 gene core promoter (CP) (boxed) with variants reported here (spanning nucleotides −239 to −187, see text) and elsewhere (*) [Citation4], CTCF (boxed), and putative transcription factor binding elements. RB1 and LINC00441 transcription start sites (arrowed). Transcription factor pRB (grey ellipse) known to bind E2F directly.
![Figure 2. (a) Exons, 5ʹUTR, CTCF binding site, Core promoter region and long non-coding RNA exons. (b) CTCF binding site (boxed, contact regions in black), RB1 gene core promoter (CP) (boxed) with variants reported here (spanning nucleotides −239 to −187, see text) and elsewhere (*) [Citation4], CTCF (boxed), and putative transcription factor binding elements. RB1 and LINC00441 transcription start sites (arrowed). Transcription factor pRB (grey ellipse) known to bind E2F directly.](/cms/asset/ace25a96-1930-495d-ba54-3fe4caf901ec/kepi_a_1834911_f0002_b.gif)
Figure 3. RB1 promoter CpG methylation profile of a Pr−/- retinoblastoma (1706) homozygous for the promoter variant c.-212 G > T spanning CpGs −627-+77 (underlined). (a) pie chart summary of RB1 promoter DNA methylation of CpGs that include putative CTCF binding site, core promoter (CP), coding and untranslated exons; (b) bar chart summary of 1706 CpG methylation of RB1 upstream region −627 to+77; (c) bar chart summary of normal peripheral blood DNA.
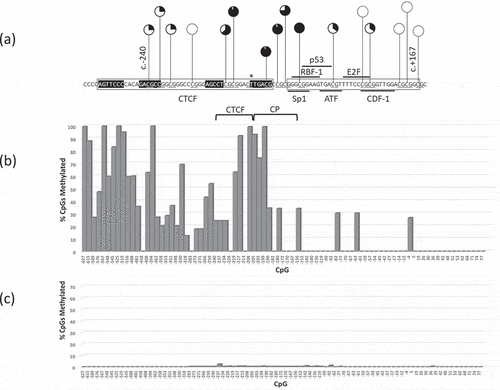
Figure 4. Bar chart summary of RB1 promoter CpG methylation profiles (CpGs c.-627-+77) of (a) 3208 (b) 3532 and (c) M22507; (a) and (b) are retinoblastomas homozygous for the RB1 variant c.-198 G > A, (c) retinoblastoma homozygous for c.-197_-196delinsTT variant.
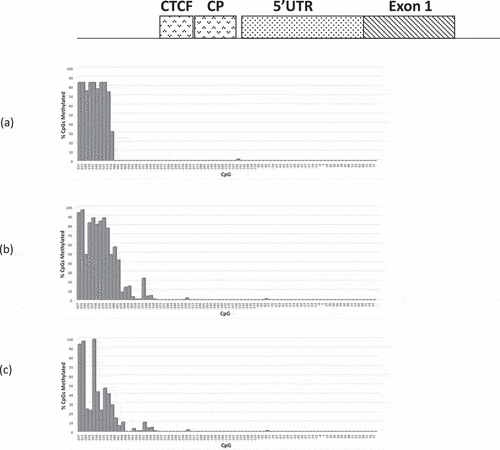
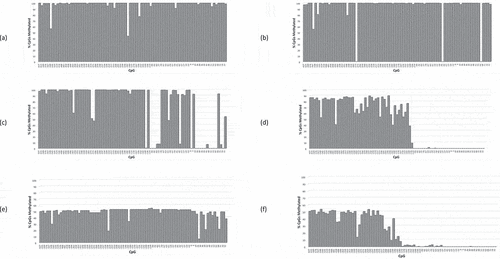
Figure 5. RB1 promoter CpG methylation profiles (CpGs −627 – +77) of PrE-/E- retinoblastomas, patients (a) 5492, (b) 3093, (c) 4927, and (d) 2504; and PrE-/E+ retinoblastomas, patients (e) 17,921 and (f) 2958.