Figures & data
Figure 1. Identification of non-templated additions (NTAs) of miRNAs
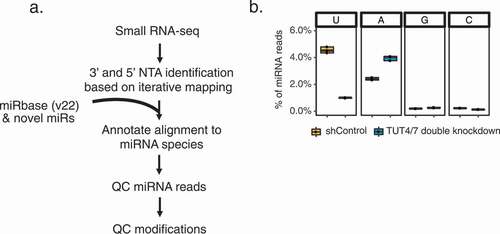
Figure 2. Generation of a comprehensive catalogue of extracellular miRNAs
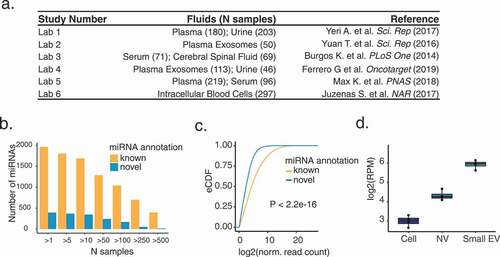
Figure 3. NTA profiles of extracellular miRNAs across biofluids
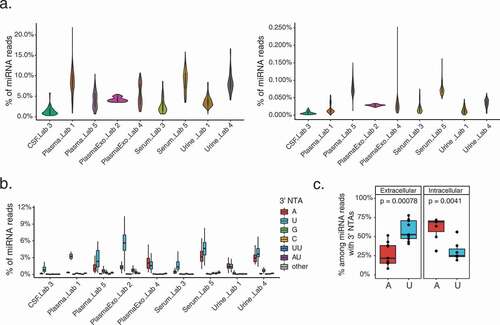
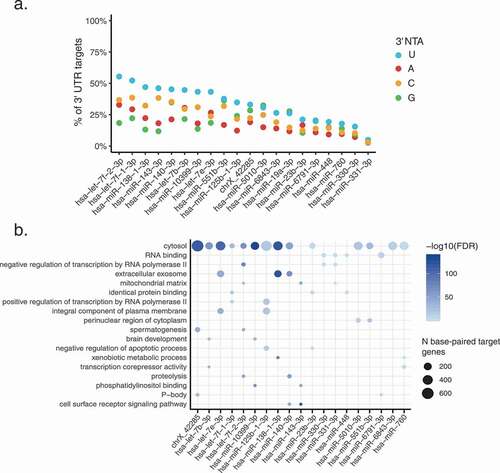
Figure 4. Characteristics of miRNAs with 3’ NTAs
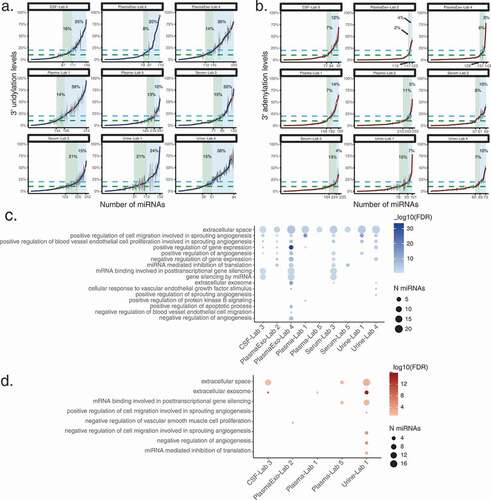
Figure 5. Distinct miRNA 3’ uridylation profiles across fluids
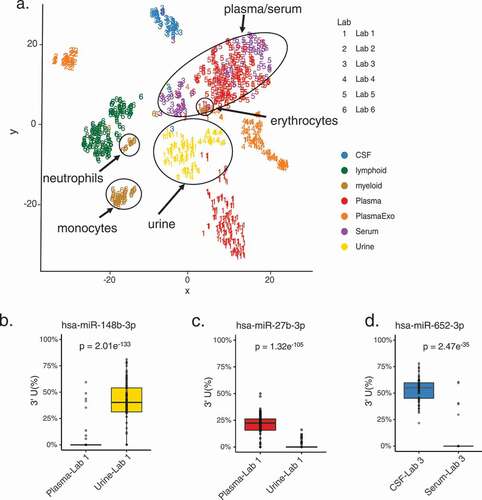
Figure 6. 3’ U base-pairs with predicted miRNA targets more often than other 3’ NTAs