Figures & data
Figure 1. Reduced MethylCap protein improves low-input methylation enrichment.
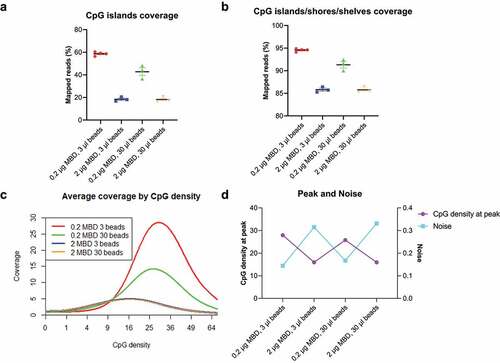
Figure 2. Methylated filler DNA is needed to compensate for low-input methylation enrichment.
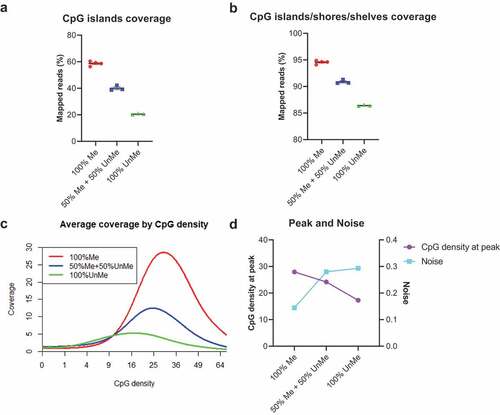
Figure 3. Different input DNA amounts in cfMBD-seq.
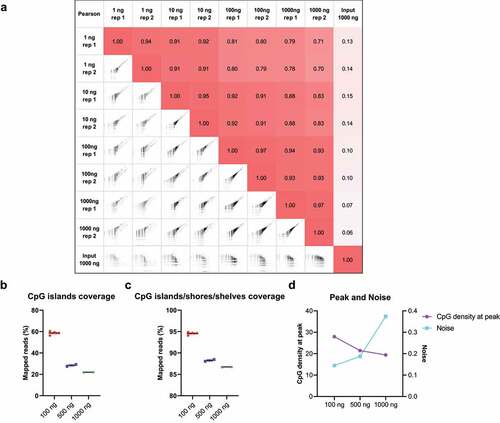
Figure 4. Comparison of cfMBD-seq with low input MBD-seq and cfMeDIP-seq.
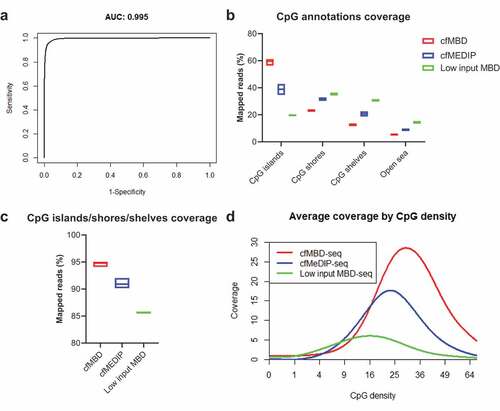
Table 1. Feature comparison among cfMBD-seq, low input MBD-seq, and cfMeDIP-seq
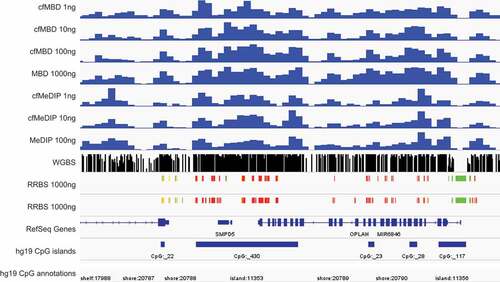
Figure 5. cfMBD-seq recapitulates methylation profiles from other technologies.
Supplemental Material
Download Zip (1.8 MB)Availability of materials and data
The sequencing data is available from GEO under the accession number GSE161331.
The data analysis pipeline is available at https://github.com/LiangWangLab/cfMBD-seq.
