Figures & data
Table 1. Clinical characteristics of the CF twins.
Table 2. Clinical characteristics of lung cancer cases and healthy controls individually matched by age, gender, smoking status and packyears.
Figure 1. Blood cell-type contribution in CF twins based on whole blood DNA methylation. (a) Estimated cell-type distribution (%) of six different cell-types in peripheral blood according to DNA methylation values in the 44 subjects representing 22 twin pairs. Heatmap of the contributions of six different cell-types in peripheral blood. Twin pairs are separated by black vertical lines. The twin with the milder phenotype is displayed on the left, whereas the one with the more severe phenotype is shown on the right. (b) Scatterplot of intra-pair cell-type contribution differences for six different cell-types. Clinical intra-pair discordance is indicated in colour.
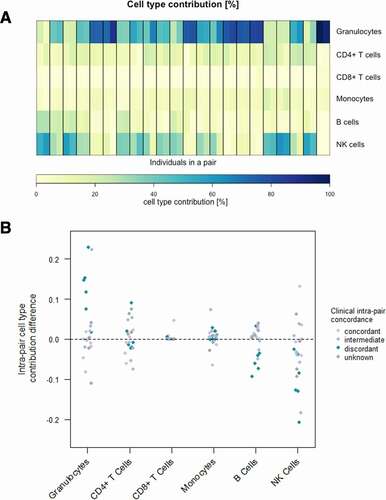
Figure 2. Selection process applied for identification of acquired inflammation-related differentially methylated positions (iDMPs). 75.2% of all probes measured passed a strict quality control (QC) including removal of cross-reactive and unreliable probes, probes with missing values and probes located in repetitive sequences. 0.2% of all retained probes are considered disease-related differentially methylated according to the assumptions described in the methods. Given that 87% of iDMPs are significantly differentially methylated in different blood cell-types, 106 probes are presumed to be acquired iDMPs.
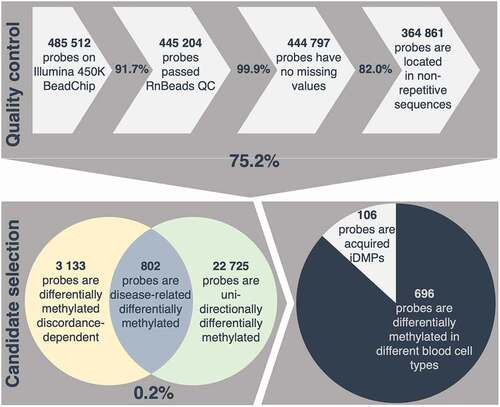
Figure 3. Methylation and intra-pair methylation differences in CF twins. (a) Absolute intra-pair methylation differences of putative acquired iDMPs. Clinical discordance is indicated in colour. Methylation differences are significantly higher (p = 0.004, one-sided Wilcoxon test) in discordant than in concordant twin pairs. The means and the medians of the absolute sums of the differences increase with the extent of clinical discordance. (b) Heatmap of methylation [%] in candidate iDMPs in 44 individuals. Rows and columns are clustered by complete hierarchical clustering and Euclidean distance.
![Figure 3. Methylation and intra-pair methylation differences in CF twins. (a) Absolute intra-pair methylation differences of putative acquired iDMPs. Clinical discordance is indicated in colour. Methylation differences are significantly higher (p = 0.004, one-sided Wilcoxon test) in discordant than in concordant twin pairs. The means and the medians of the absolute sums of the differences increase with the extent of clinical discordance. (b) Heatmap of methylation [%] in candidate iDMPs in 44 individuals. Rows and columns are clustered by complete hierarchical clustering and Euclidean distance.](/cms/asset/11b1fc26-8fa0-4d44-bb03-b5c5999c07f3/kepi_a_1959976_f0003_oc.jpg)
Figure 4. DNA methylation differences in lung cancer cases compared to healthy controls. (a) Heatmap of methylation differences in CG dinucleotides of 20 genomic regions (numbers), corresponding to 21 iDMPs (rows separated by black lines) in 209 pairs (columns). NAs are shown in black, clinical parameters are colour coded (b) Bar plot of the means of all absolute intra-pair methylation differences [%] per CG dinucleotide of the 20 numbered regions. Median values across the regions are indicated. The width of the boxes represents the number of observations (i.e., the number of CG dinucleotides per region).
![Figure 4. DNA methylation differences in lung cancer cases compared to healthy controls. (a) Heatmap of methylation differences in CG dinucleotides of 20 genomic regions (numbers), corresponding to 21 iDMPs (rows separated by black lines) in 209 pairs (columns). NAs are shown in black, clinical parameters are colour coded (b) Bar plot of the means of all absolute intra-pair methylation differences [%] per CG dinucleotide of the 20 numbered regions. Median values across the regions are indicated. The width of the boxes represents the number of observations (i.e., the number of CG dinucleotides per region).](/cms/asset/35741e0e-da13-4234-a7e5-dbc4a8d13277/kepi_a_1959976_f0004_oc.jpg)
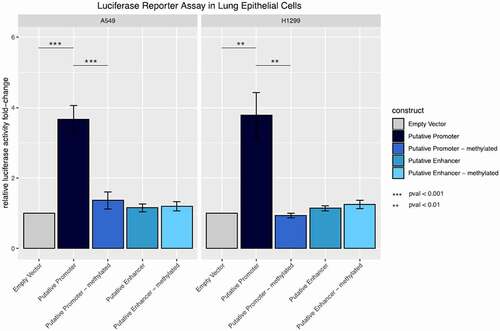
Figure 5. Promoter and enhancer activity of the methylated and unmethylated inflammation-related DMR in lung epithelial cells. Fold change relative luciferase activity in A549 and H1299 cells. The empty vector corresponds to a construct without promoter sequence. For the putative promoter vector, the sequence of interest has been cloned into the empty vector upstream of the luciferase gene. For the putative enhancer construct, the region of interest was cloned upstream of a minimal promoter for the luciferase gene. Both vectors were compared to their in silico methylated counterparts.