Figures & data
Table 1. Demographic characteristics and benzene occupational exposure level.
Table 2. Demographic characteristics and formaldehyde occupational exposure level.
Table 3. Genome-wide significant (Bonferroni corrected P-value <0.05) differentially mean methylated probes (DMPs) from a cross-sectional study of occupational exposure to benzene, where differential methylation was assessed with respect to continuous benzene exposure.
Table 4. Top 25 out of 318 genome-wide significant (Bonferroni corrected P-value <0.05) differentially methylated probes, where differential methylation was defined in terms of the variance of CpG probe methylation (DVPs), from a cross-sectional study of occupational exposure to benzene, and differential methylation was assessed with respect to continuous benzene exposure.
Table 5. Genome-wide significant (Bonferroni corrected P-value <0.05) differentially variable probes (DVPs) from a cross-sectional study of occupational exposure to formaldehyde, where differential methylation was assessed with respect to binary, exposed versus controls, comparison groups.
Figure 1. Formaldehyde-associated differential DNA methylation in the DUSP22 gene, highlighting nine differentially variable probes (DVPs) that exhibited significant (Bonferroni-adjusted P-values <0.05) decreased variance of DNA methylation in formaldehyde-exposed workers compared to controls (a). All CpG probes on the 450 K array that are annotated to the DUSP22 gene (excluding any that might have been filtered out) are shown, with the nine significant DVPs being those in the red box (A). The pronounced significance of these 9 DVPs can be seen in the Manhattan plot, where the red and blue lines represent the family-wise error rate (FWER) and false discovery rate (FDR) thresholds, respectively, for statistical significance (b).
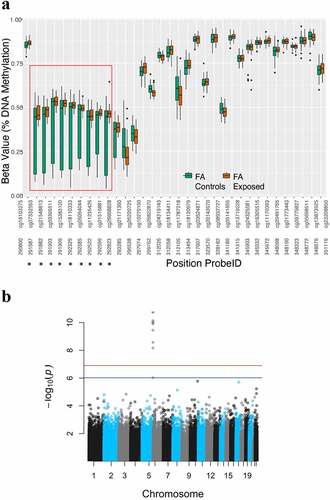
Figure 2. Genomic annotation of two regions exhibiting differential mean methylation (DMRs) in formaldehyde-exposed subjects versus controls. A 2,174 base-pair region on chromosome 7, comprised of 44 CpG probes, and located in the promoter of HOXA5 exhibited significant hypomethylation in formaldehyde exposed workers compared to controls, with a family-wise error rate (FWER)-controlled P-value = 0.01 (a). A 1,598 base-pair region on chromosome 6, comprised of 10 CpG probes, and located in the DUSP22 gene exhibited nearly genome-wide significant hypermethylation in formaldehyde exposed subjects compared to controls, with an FWER-controlled P-value = 0.063 (b).

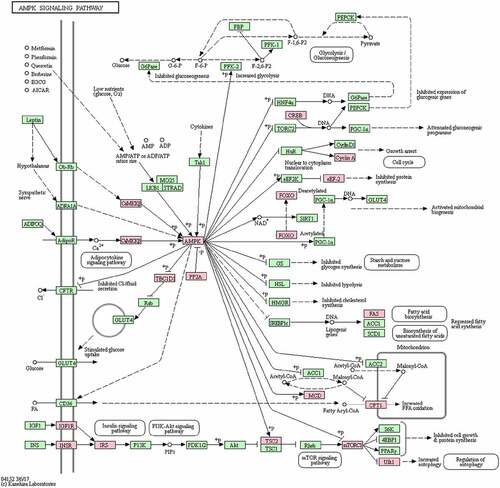
Figure 3. Benzene occupational exposure-associated enrichment of the AMP-activated protein kinase (AMPK) signalling pathway. This was the most significant finding from the gene set enrichment analysis (GSEA) of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The 20 genes that corresponded to sites exhibiting significant, benzene exposure-associated differential DNA methylation are highlighted in pink.
Supplemental Material
Download Zip (852.1 KB)Data availability statement
Summary results and analysis code (R) for both EWAS are available on GitHub (https://github.com/rachaelvp/EWAS-BZ-FA) and Open Science Framework (https://osf.io/exqzy/).
