Figures & data
Figure 1. circDHRS3 is down-regulated in high-grade prostate cancer. (a). Cluster heatmap for circRNAs among various pathological diagnoses (filtered by p < 0.05 and |log2FC| > 1). Columns show tissues while rows show differentially expressed circRNAs, with circDHRS3 identified on the right of rows. (b). qRT-PCR was used to look at the expression of circDHRS3 in normal prostate epithelial cells RWPE-1 and prostate cancer cell lines 22Rv1, PC3, and Du145. The data were presented as means ± SD. (***p < 0.001, **p < 0.01 versus RWPE-1). (c). Expression of circDHRS3 and linear mRNA-DHRS3 either treated or left untreated with RNase R as detected by qRT-PCR. circDHRS3 is more resistant to RNase R than mRNA. Data are presented as the mean ± SD. (d). qRT-PCR detected the residual level of circDHRS3 and linear mRNA-DHRS3 in Du145 cells treated with Actinomycin D at the specific time point. Data were shown as the means ± SD. (e). Tissue microarray data in prostate cancer tissues compared to precancerous prostate tissues by ISH revealed that circDHRS3 is downregulated in prostate cancer tissues (Left panel) compared to paracancerous prostate tissue (Right panel). (f). Survival curves of patients with prostate cancer of high or low expression of circDHRS3, according to the ISH, using the median value as the grouping criterion. The follow-up time spans 7 years after surgery.
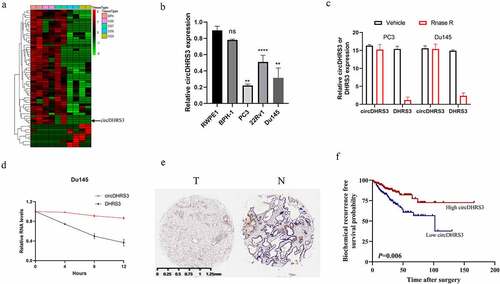
Figure 2. Overexpression of circDHRS3 decreased the proliferation and metastasis of prostate cancer cell lines in vitro. (a). qRT-PCR was used to determine the efficacy of LV-circDHRS3 overexpression in Du145 and PC3 cells, compared to the negative control (NC). Data are presented as the mean ± SD. (b). CCK-8 proliferation assays of Du145 and PC3 were detected to perform the proliferation of LV-circDHRS3 and NC. Data are presented as the mean ± SD. (c). A colony formation experiment was used to determine the ability of Du145 and PC3 cells transfected with LV-circDHRS3 and LV-NC to produce colonies. (d). The proliferation ability of Du145 and PC3 cells transfected with LV-circDHRS3, and LV-NC was assessed using an EdU assay. (Scale bars = 50 μm). (e). Invasion ability of Du145 and PC3 cells transfected with LV-circDHRS3, and LV-NC was determined using transwell assays. (Scale bars = 100 μm). (f). Wound healing was applied to assess the migration ability of Du145 and PC3 cells transfected with LV-circDHRS3 and LV-NC.
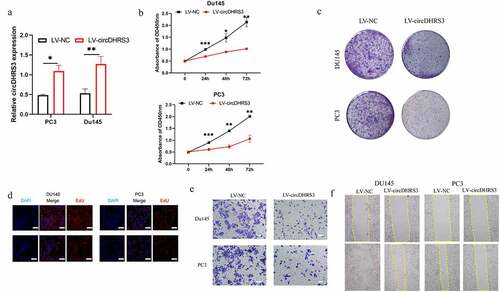
Figure 3. circDHRS3 inhibits the expression of miRNA and serves as an RNA sponge for miR-421 and miR-421 serves as an mRNA inhibitor to downregulate MEIS2 expression. (a and b). Venn diagram of the predicted downstream targets of circDHRS3 and upstream targets of MEIS2 from Encori and software circMIR, shown as a schematic illustration. (c). The affinity of circDHRS3 for miR-421 in PC3 cells was assessed using RNA pull-down. qRT-PCR detection of relative miRNA expression under the enriched circDHRS3 probe. Data are presented as mean ± SD. (d). RIP experiments were performed in PC3 cells against anti-immunoglobulin G or Ago2, and the precipitated circDHRS3 and miR-421 were detected by qRT-PCR. (e). FISH showed the subcellular co-localization between circDHRS3(red) and miR-421(green). DAPI (blue) was used to stain the nucleus. (Scale bars = 25 μm). (f). Dual-Luc reporter assays show the binding properties of circDHRS3 and miR-421. The Mut version of circDHRS3 is also shown. (g). The GEPIA database showed MEIS2 down-expression in prostate cancer tissue versus normal tissue. *p < 0.05. (h). Dual-Luc reporter assays showed binding properties of miR-421 and MEIS2. The Mut version of the 3ʹUTRMEIS2 is also shown. (i). Relative Luc activity was determined 2 days after transfection with the miR-421 mimic/normal control or with the 3ʹUTR-MEIS2 WT/Mut in PC3 cells. Data are presented as the mean ± SD. (j). Western blot displayed the MEIS2 levels among PC3 and Du145 cells, both of which were transfected with miR-NC, miR-mimic, miR-inhibitor-NC, or miR-inhibitor, respectively. GAPDH was used as the reference gene.
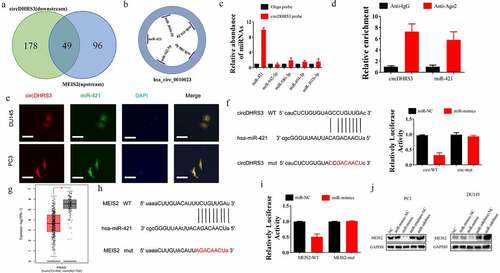
Figure 4. miR-421 overexpression or MEIS2 silencing restored proliferation, migration, and invasion after circDHRS3 overexpression. (a and b). Colony formation assays were performed to detect the colony formation ability of Du145 and PC3 cells transfected with LV-NC, LV-circDHRS3, LV-circDHRS3+ miR-mimic, LV-circDHRS3+ sh-MEIS2, miR-inhibitor, and miR-inhibitor + sh-MEIS2. Data are presented as the mean ± SD. (c and d). Transwell assays were performed to detect the colony formation ability of Du145 and PC3 cells transfected with LV-NC, LV-circDHRS3, LV-circDHRS3+ miR-mimic, LV-circDHRS3+ sh-MEIS2, miR-inhibitor, and miR-inhibitor + sh-MEIS2. Data are presented as the mean ± SD. (Scale bars = 100 μm).
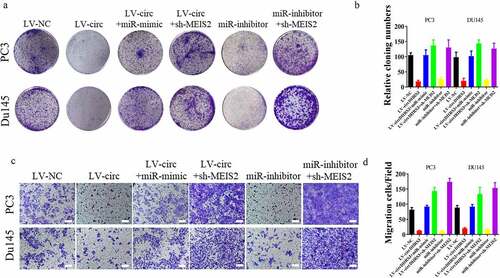
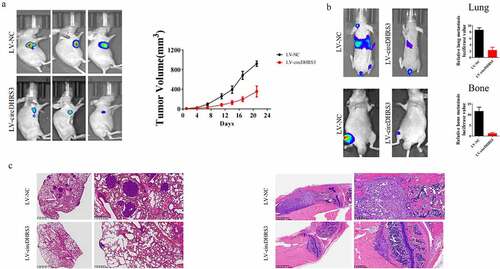
Figure 6. Overexpression of circDHRS3 decreased prostate cancer proliferation and metastases in vivo. (a). Tumour sizes were measured and compared for LV-NC-PC3 and LV-circDHRS3-PC3 xenografts in nude mice after 21 days. Tumour formation was detected using live imaging. (b). The lung or bone metastasis ability of LV-NC-PC3-Luc and LV-circDHRS3-Luc by intravenous tail injection or intratibial injection was detected using live imaging. (c). HE staining of lung and bone after injection of LV-NC-PC3-Luc and LV-circDHRS3-Luc.
Supplemental Material
Download Zip (12.7 MB)Data availability statement
On reasonable request, the corresponding author can provide the datasets used and/or analyzed during this investigation. The data that support the findings of this study are available from the corresponding author, Haowen Jiang([email protected]), upon reasonable request.