Figures & data
Figure 1. Hsa_circ_0004680 is up-regulated and correlated with poor clinical outcomes in HCC patients. (a) The difference of expression in six paired of HCC samples. (b) High and low expression of circle RNAs in HCC tissue. (c) 315 circle RNAs were high expressed in HCC. (d) circ_0004680 was high expression in HCC. (e) HCC tissue array was stained by RNA in situ hybridization. (f) High expression of circ-CCT3 showed poor prognosis in HCC patients, p = 0.0011. (g)Relationships between clinicopathology (including pathological grade, AFP level, and TNM stage) and circ_004680 in HCC tissues. The results revealed that the mRNA level of circ_004680 was correlated with pathological grade, AFP level and TNM classification.
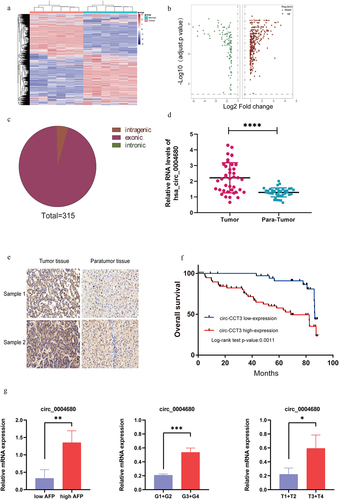
Figure 2. Characteristics of hsa_circ_0004680.(a) Sanger sequencing of RT-PCR products performed by circ-CCT3 primer. (b) circ_0004680 derived from exon 3,4 and 5 of gene CCT3. (c) circ-CCT3 can not be digested by RNase R, while it’s parental gene CCT3 was seriously digested.***p < 0.001. (d) circ-CCT3 can be amplified by random hexamer primer. (e) circ-CCT3 was restrained by Actinomycin D. (f) PCR results showed circ-CCT3 was mainly located in cytoplasm. (g) the electrophoresis experiments result of circ-CCT3 in LM3 and Huh7 cell lines.
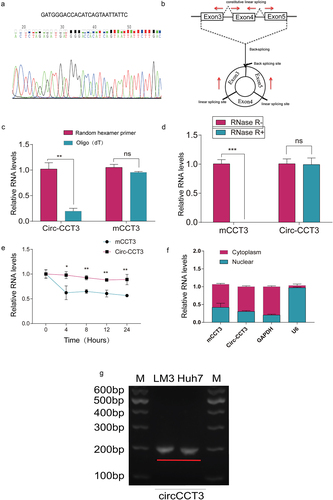
Figure 3. Knock-down of circ-CCT3 suppresses the proliferation of HCC cell lines both in vitro and in vivo. (a) CCK8 assay indicated that knock-down circ-CCT3 inhibited the proliferation of HCC-LM3 and Huh7 cells. (b) EDU assay showed that knock-down of circ-CCT3 inhibited the proliferation of HCC-LM3 and Huh7 cells. (c) Colony formation assay showed knock-down of circ-CCT3 restrained the growth of HCC-LM3 and Huh7 cells. (d) Cell cycle analysis showed knock-down of circ-CCT3 arrested HCC-LM3 and Huh7 cells G2 phase. (e) Transwell assay indicated that knock-down of circ-CCT3 inhibited the invasion and metastasis of HCC-LM3 and Huh7 cells in vitro. (f) Knock-down of circ-CCT3 restrained the growth of xenograft mice tumour.(g) Knock-down of circ-CCT3 inhibited lung metastasis in nude mice..
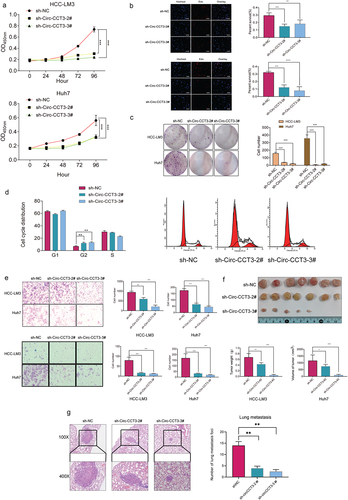
Figure 4. Downregulation of circ-CCT3 plays a tumour suppression role in HCC via the miR-378a-3p-FLT-1pathway. (a) the 11 potential target genes of miR-378-3p. (b) Luciferase reporter assays were performed with both WT and Mut plasmids. (c, d) Expression of FLT1 following circ-CCT3 overexpression or knock-down was evaluated by RT-Qpcr and western blotting in HCC cell lines. (e) Rescue experiments were performed to confirm the regulation of FLT-1 was under the control of circ-CCT3 and miR-378a-3p. (f) Tube formation assay was performed to evaluate the angiogenesis after knock-down of circ-CCT3 in HUVECs. (g) Transwell assay was conducted to evaluate the metastasis of HCC cells after overexpressing circ-CCT3.
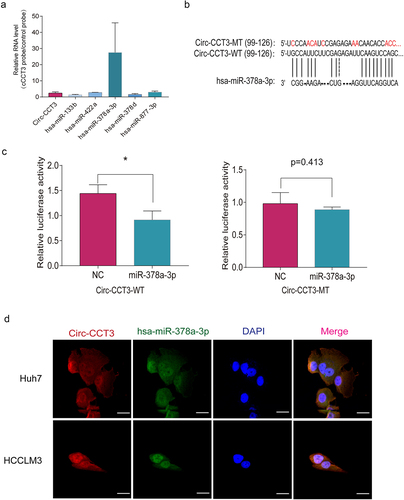
Figure 5. Circ-CCT3 serves as sponge for miR-378a-3p. (a) RNA immunoprecipitation assay showed circ-CCT3 interacted with miR-378a-3p.(b) Examples of the potential bindings between circ-CCT3 conserved sequence and miR-378a-3p. (c) Dual-Luciferase assay showed miR-378a-3p combined with wild type circ-CCT3 .(d) Dual-Luciferase assay showed miR-378a-3p combined with mutant type circ-CCT3. (e) Co-localization between miR-378-3p and circ-CCT3 observed by RNA in situ hybridization in Huh7 and HCC-LM3 cells. Nuclei were stained with DAPI.
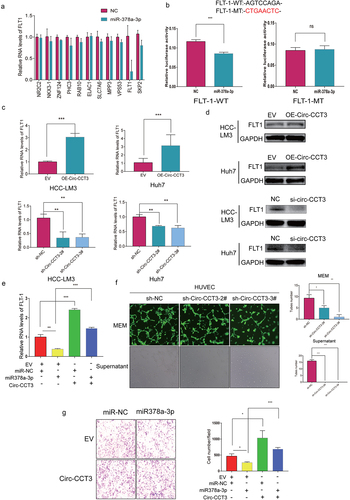
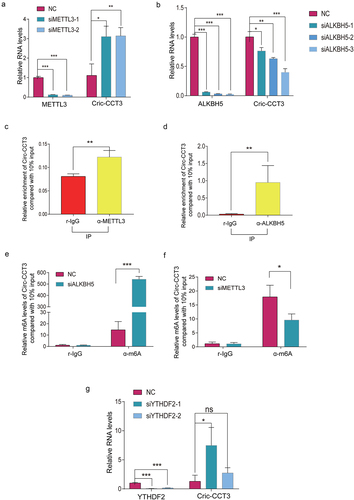
Figure 6. Circ-CCT3 is modified by m6A . (a) Knock-down of METTL3 increased circ-CCT3 level. (b) Knock-down of ALKBH5 decreased circ-CCT3 level. (c, d) RNA pull-down showed METTL3 and ALKBH5 combined with circ-CCT3. (e) Knock-down of ALKBH5 increased circ-CCT3 m6A level. (f) Knock-down of METTL3 decreased circ-CCT3 m6A level. (g) Knock-down of YTHDF2 increased circ-CCT3 m6A level.
Supplemental Material
Download Zip (4.4 MB)Data availability statement
10.57760/sciencedb.j00001.00512