Figures & data
Table 1. The identity and similarity of N-terminus tail in histone subunits H3.1, H3.3, and H4 of Arabidopsis thaliana across organisms.
Figure 1. Comparison of PRC2 core components and their associated factors among Drosophila, human, mouse, and Arabidopsis. the interaction of PRC2 core components is represented among Drosophila, human, and Arabidopsis. The core components used the same colour between the organisms indicate corresponding orthologous proteins. The associated factors are classified by their functions, and their colour-coding indicates orthologous proteins between organisms.
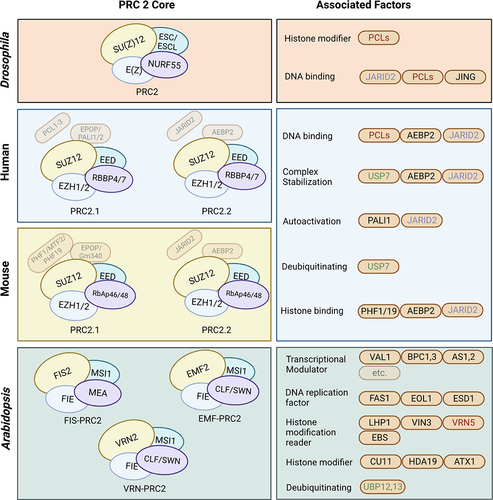
Figure 2. Comparison of COMPASS core components and their associated factors among Drosophila, mouse, human, and Arabidopsis. The interaction of COMPASS core components is represented among Drosophila, mouse, human, and Arabidopsis. The core components used the same colour between the organisms indicate corresponding orthologous proteins. The associated factors are classified by their functions, and their colour-coding indicates orthologous proteins between organisms.
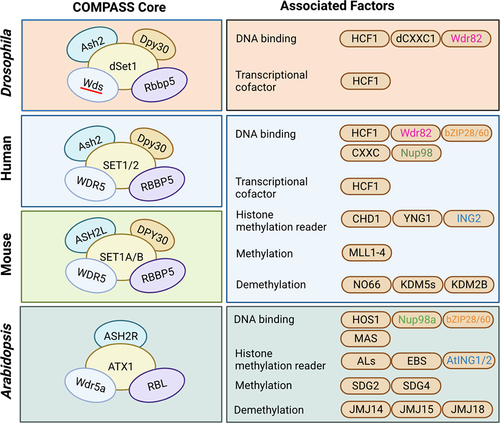
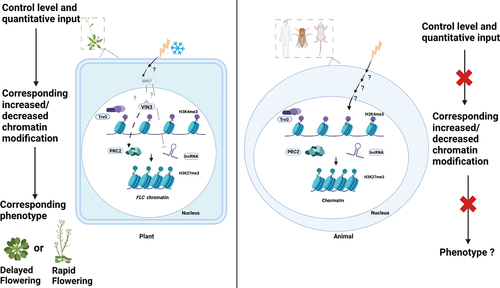
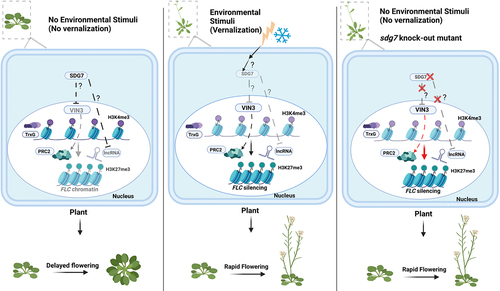
Figure 3. Vernalization pathway without or with prolonged cold exposure as an environmental stimulus in Arabidopsis. Quantitative correlations between the duration of cold exposure and the histone modifications results in changes of FLC chromatin status and its expression. Because the flowering time in vernalization pathway depends on FLC expression, the flowering time is switched based on the quantitative environmental input (left and middle panel). The sdg7 mutant is unable to suppress the expression of the PRC2 component VIN3, as well as the lncRNAs COOLAIR and COLDAIR. This results in a decrease in FLC expression via an increase in H3K27me3, leading to rapid flowering without cold exposure (right panel).
Supplemental Material
Download Zip (2.7 MB)Data availability statement
histone sequences used in this study are available at the UniProt (https://www.uniprot.org/) [Citation265] and NCBI RefSeq (https://www.ncbi.nlm.nih.gov/refseq/) [Citation266]. Accession numbers are listed in Supplement .