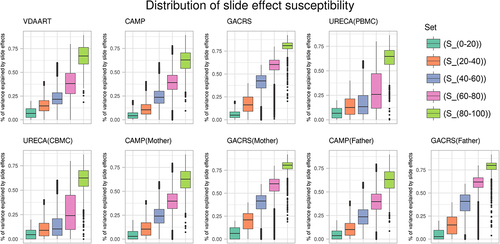
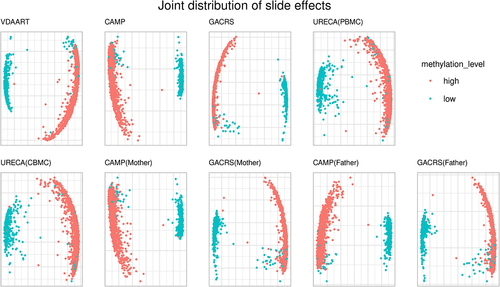
Figures & data
Supplemental Material
Download Zip (14.8 MB)Data availability statement
GACRS and CAMP: All TOPMed data can be requested for access and can be made available through the TOPMed consortium after careful review and approval by the TOPMed Data Access Committee (https://topmed.nhlbi.nih.gov/). Participant consent and Data Use Limitations differ within and across TOPMed studies and should be requested individually. Additional documentation, such as local IRB approval and/or letters of collaboration with the primary study PI(s) may be required. DNA methylation data for the URECA cohort is available in the Gene Expression Omnibus (GEO) repository under the accession number GSE132181 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132181).
EWAS atlas: https://ngdc.cncb.ac.cn/ewas/atlas.
EWAS Toolkit: https://ngdc.cncb.ac.cn/ewas/toolkit.
Illumina EPIC v1.0 support files: https://support.illumina.com/downloads/infinium-methylationepic-v1-0-product-files.html.
Illumina EPIC v2.0 support files: https://emea.support.illumina.com/downloads/infinium-methylationepic-v2-0-product-files.html.
R statistical software, version 4.1.0: https://www.r-project.org/.
cgageR R package: https://rdrr.io/github/metamaden/cgageR/.