Figures & data
Figure 1. Synopsis of the experience.
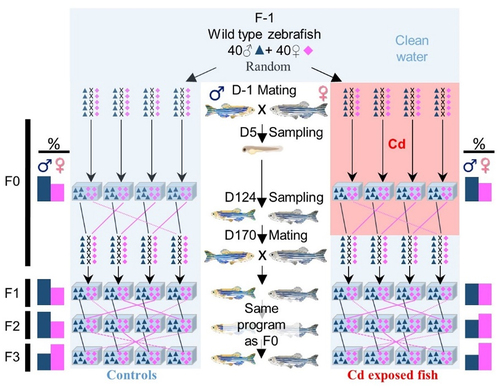
Figure 2. Cd exposure induced changes in allele frequencies.
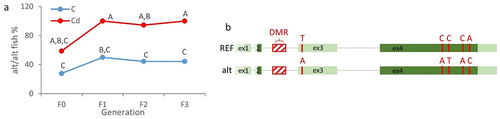
Figure 3. The exon 3 of zbtb38 is transcribed independently of the other exons.
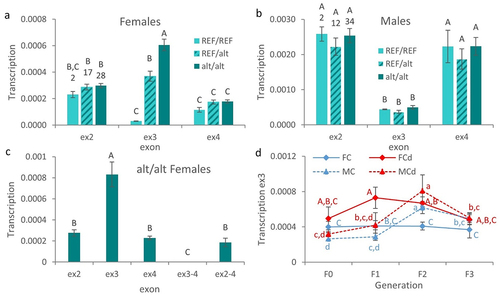
Figure 4. Sex-specific patterns of DNA methylation and RNA transcription.
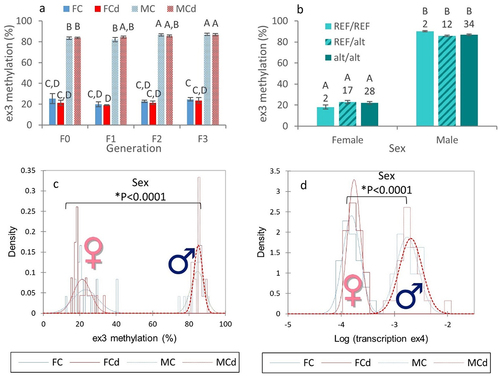
Figure 5. A complex dialog between the environment, the genome and DNA methylation led to variations in the transcription level of the ex3-NAT of zbtb38.
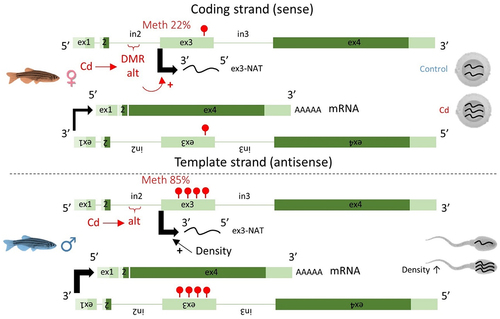
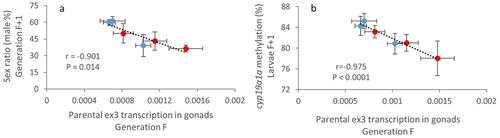
Figure 6. Parental expression of ex3-NAT influences offspring sex ratio.
Supplemental Material
Download MS Word (721 KB)Data availability statement
Raw sequencing data were deposited in the European Nucleotide Archive with accession number PRJEB52137. All other data are included in the article and/or supporting information.
