Figures & data
Figure 1. Schematic of scCuT&Tag applied to cultured cells and tissues in different species. Cells or nuclei suspension are isolated from cell culture, mice tissues, human tissues and plant tissues. These cells or nuclei are subjected to antibody incubation, pA-Tn5 binding and tagmentation, then they are combined with nanowell, 10× Genomics or split-pool system to distinguish cellular identity.
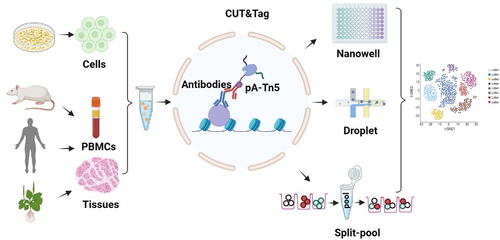
Figure 2. Multifactorial CUT&Tag strategies. (left) CUT&Tag 2 for 1 involves the use of two antibodies against H3K27me3 (heterochromatin marks) and Ser5P-RNAPII (active chromatin marks). This approach follows the same workflow as regular CUT&Tag, except for mixing two antibodies into the cells. The difference in fragment size and feature breadth (broad domains for H3K27me3 and narrow peak of Ser5P-RNAPII) are used together to calculate the deconvolution of two signals. (middle) in MuTi-Tag approach, each antibody is pre-incubated with the barcoded pA-Tn5, thus marking several different histone modifications and TFs in the same experiment. (right) nano-CUT&Tag is the use of nanobodies, single-chain antibodies directly fused to Tn5, where higher efficiency can be obtained without protein A.
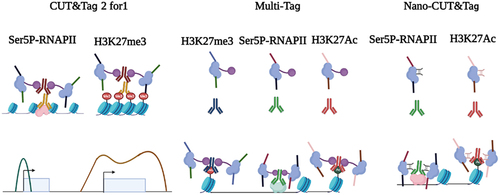
Figure 3. The principle of CUT&Tag and CUT&Tag variants. In the CUT&Tag experiment, the primary antibody recognizes the target protein and then mediates the binding of the secondary antibody and the protein A/G-Tn5 fusion protein. Tn5 can precisely target and cleave the DNA sequence near the target protein in the presence of Mg2+. Simultaneously, the sequencing adapters will be inserted into both ends of DNA fragments by Tn5. For tissue section, spatial-CUT&Tag and MERFISH can be conducted. For cell suspension, CUT&Tag and CUT&Tag variants can be performed.
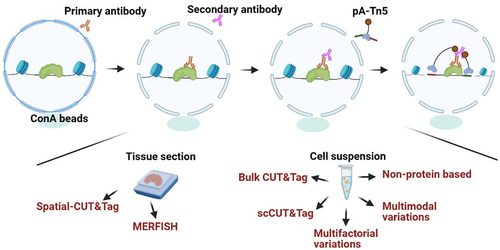
Table 1. CUT&Tag and CUT&Tag variants.
Table 2. The combinations of CUT&Tag with other omics.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.
