Figures & data
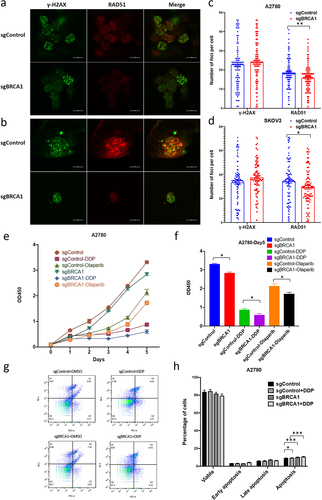
Figure 1. The relationship between methylation at CG sites within the BRCA1 promoter and gene expression levels.
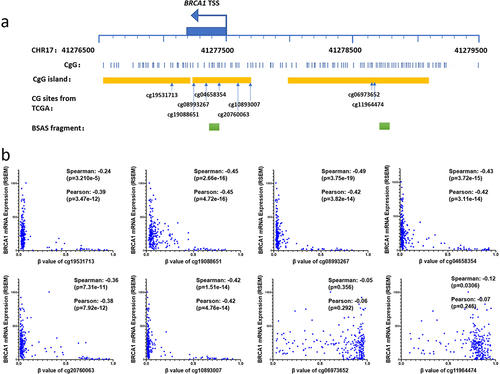
Figure 2. Methylation status of the BRCA1 promoter and its implications for OC development and homologous recombination defects (HRD).
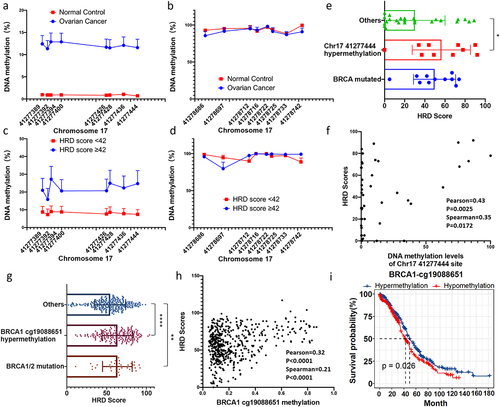
Figure 3. Design and validation of the epigenetic editor for the BRCA1 promoter.
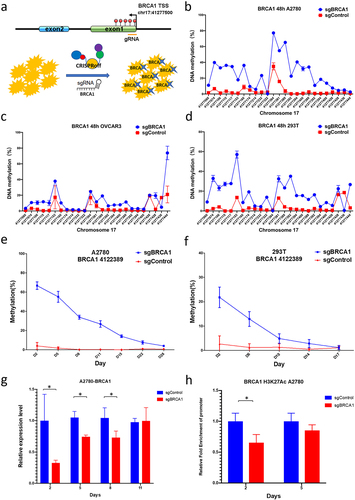
Figure 4. The impact of epigenetic editing of the BRCA1 promoter on HR repair capacity and drug sensitivity in OC cells.