Figures & data
Table 1. Study sample characteristics by chemotherapy.
Figure 1. Correlation estimates between the GrimAge surrogate biomarkers of blood plasma proteins and chemotherapy.
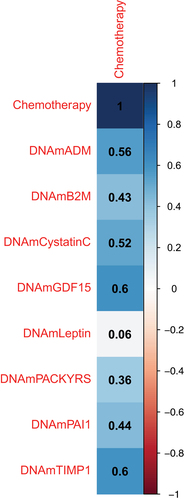
Table 2. Multiple linear regression results for the association between markers of biological ageing and chemotherapy.
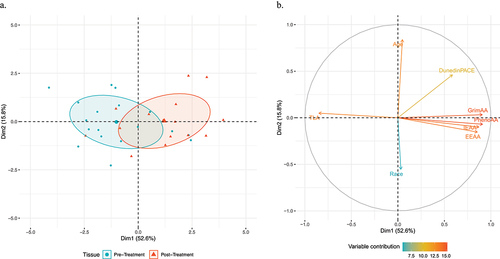
Figure 2. Principal component analysis of the association between markers of biological ageing and chemotherapy.
-) Supplemental Tables & figures.docx
Download MS Word (303.9 KB)Data availability statement
The dataset used in this analysis is available in NCBI’s Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178887, accession number GSE178887.
