Figures & data
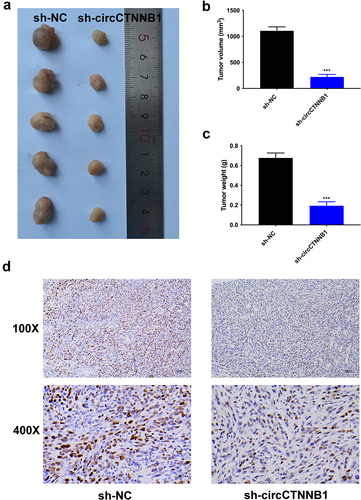
Figure 1. The higher expression of circCTNNB1 was discovered in lung cancer.
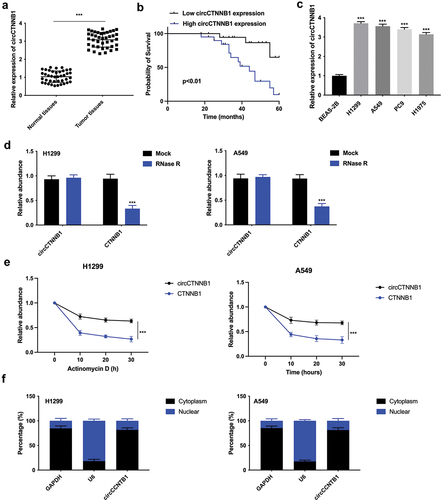
Figure 2. CircCTNNB1 facilitated lung cancer cell proliferation, migration, invasion, and suppressed cell senescence.
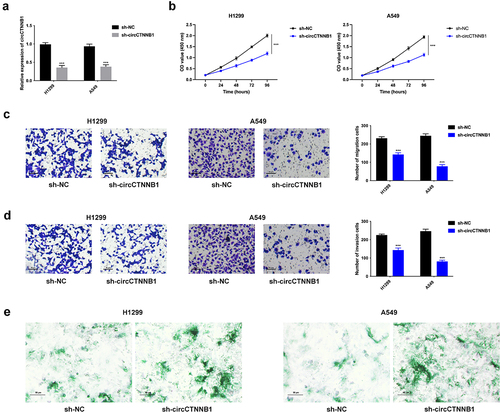
Figure 3. Knockdown of circCTNNB1 retarded the Wnt pathway.
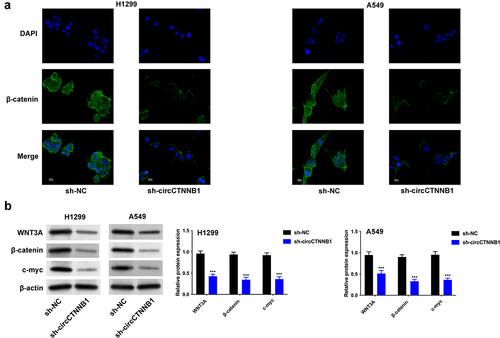
Figure 4. CircCTNNB1 combined with miR-186-5p to target YY1.
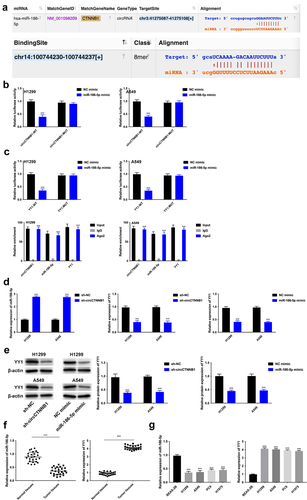
Figure 5. YY1 overexpression can rescue the decreased cell proliferation, migration, invasion, and the increased cell senescence mediated by circCTNNB1 suppression.
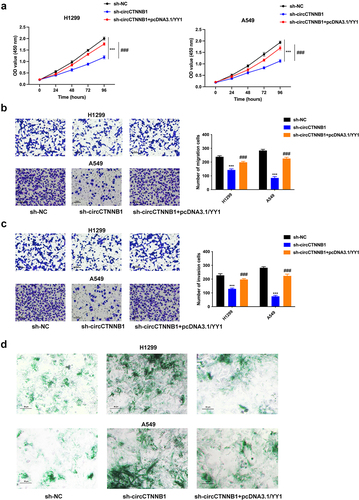
Figure 6. YY1 overexpression can rescue the retarded Wnt pathway mediated by circCTNNB1 knockdown.
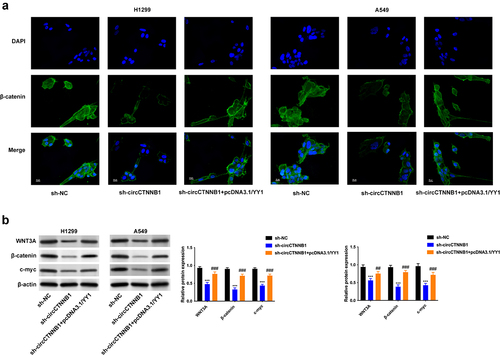
Figure 7. YY1 can transcriptionally activate circCTNNB1 to form YY1/circCTNNB1/miR-186-5p/YY1 loop.
Supplemental Material
Download TIFF Image (56 MB)Cell STR file.zip
Download Zip (2.3 MB)Data availability statement
The data that support the findings of this study are available from the corresponding author.