Figures & data
Figure 1. Tet1 mutants regulate common and distinct CpG sites in the adult cortex.
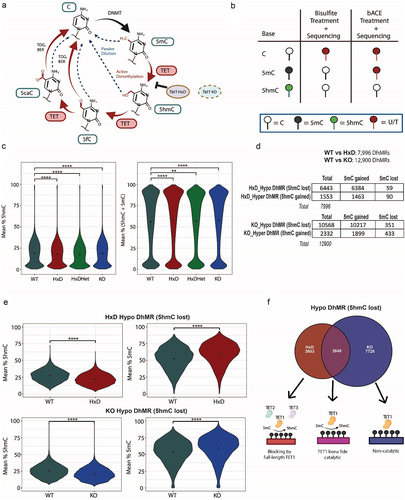
Figure 2. TET1 mainly exerts its function by modulating DhMRs in exons and introns.
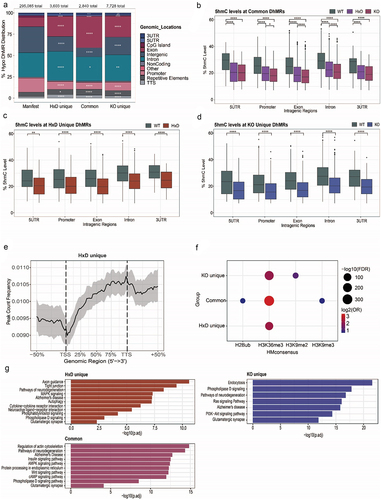
Figure 3. Tet1 perturbation is associated with decreased 5hmC.
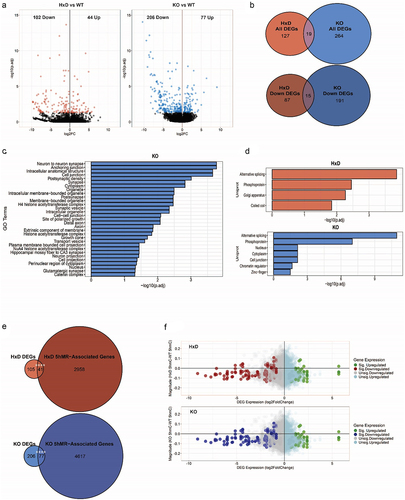
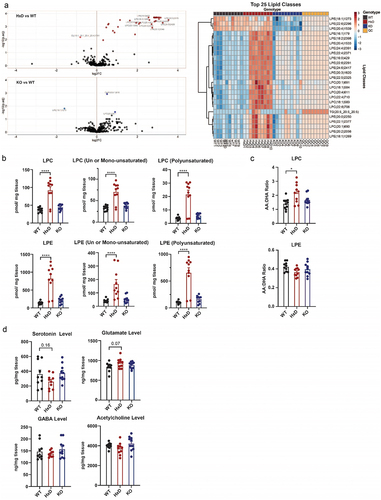
Figure 4. Tet1 HxD accumulates phospholipid LPC/LPE in the cortex while neurotransmitter levels remain intact.
Supplemental Material
Download MS Word (23.9 KB)Supplemental Material
Download MS Word (2.7 MB)TableS7_Revision.xlsx
Download MS Excel (11.7 KB)TableS6_Revision.xlsx
Download MS Excel (2.2 MB)TableS8_Revision.xlsx
Download MS Excel (42.1 KB)TableS4_Revision.xlsx
Download MS Excel (10.7 KB)TableS9_Revision.xlsx
Download MS Excel (11.1 KB)TableS1_Revision.xlsx
Download MS Excel (9 KB)TableS2_Revision.xlsx
Download MS Excel (9 KB)TableS5_Revision.xlsx
Download MS Excel (9.1 KB)TableS3_Revision.xlsx
Download MS Excel (9.6 KB)Data Availability Statement
Sequencing data have been deposited in GEO database under the accession number GSE271375 and GSE271376.
