Figures & data
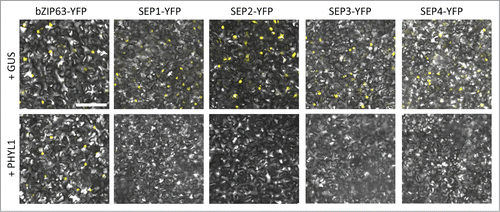
Figure 1. PHYL1 induces the degradation of SEP1–4 proteins. Agrobacterium cultures (OD600 = 1.0) for transient expression of YFP-fused proteins (bZIP63 and SEP1–4) and GUS or PHYL1 protein were mixed at a ratio of 1:10 and infiltrated into Nicotiana benthamiana leaves. The accumulation and subcellular localization of the transiently expressed YFP-fused proteins were observed 50 h after infiltration. Scale bar = 100 µm. The Agrobacterium strain and plasmids were the same as in the previous study, respectively.Citation15 The composition of the infiltration buffer is as follows: 10 mM morpholinepropanesulfonic acid (MES, pH 5.6), 10 mM MgCl2, and 150 µM acetosyringone. Nuclear localization of the transcription factors used in this study was confirmed by DAPI staining (data not shown).