Figures & data
Table 1. Sequence composition of Bd-SCL33 splice variants in Brachypodiuma
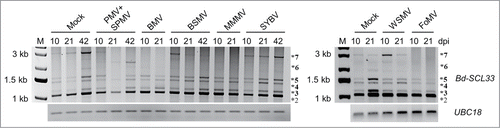
Figure 1. Bd-SCL33 differential splicing during diverse grass virus infections. Expression of Bd-SCL33 transcripts in mock, PMV+SPMV-, BMV-, BSMV-, MMMV-, SYBV-, WSMV- and FoMV-infected Brachypodium at 10, 21 and 42 days postinoculation (dpi), as determined by RT-PCR. The alternatively spliced Bd-SCL33 transcripts, as defined in , are identified by an asterisk (*). UBC18 was used as control for approximately equal cDNA amounts in the different samples. Lane “M” indicates DNA molecular weight markers.