Figures & data
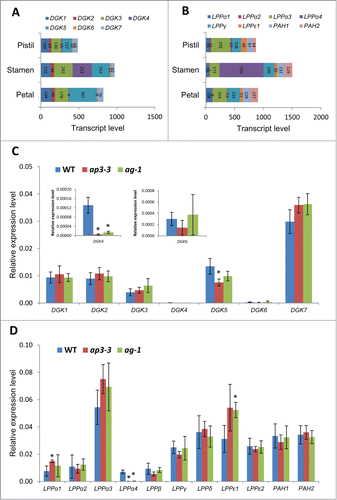
Figure 1. Phosphatidic acid (PA) profiles of the flowers of ap3-3 and ag-1 compared to the wild type analyzed by triple quadrupole LC-MS/MS. (A) Levels of major PA molecular species. (B) Levels of major PC molecular species. (C) Levels of total PA and PC. Data are mean ±SD of 4 biological replicates. Asterisks indicate significance (p < 0.01) from wild type. Mature flowers of wild type or homeotic mutants (ap3-3 and ag-1) were harvested, immediately frozen in liquid nitrogen, and kept at −80°C until lipid extraction. Prior to lipid extraction, frozen tissues were incubated in hot (75°C) isopropanol containing 0.05% (v/v) butylated hydroxytoluene (Cat. No. B1378, Sigma-Aldrich, St. Louis, MO) for 15 min to inhibit phospholipase D activity for PA production. PA and phosphatidylcholine (PC) were analyzed as follows; the dried lipid fractions extracted from plant tissues were dissolved in 200 μl chloroform/methanol 1/1 (v/v). Samples were then transferred (10 μl) into MS vials and mixed with an equal volume of a 0.2 μg/ml solution of internal standards dimyristoylphosphatidic acid (PA 14:0/14:0, 0.32 nmol/ml) and dimyristoylphosphatidylcholine (PC 14:0/14:0, 0.30 nmol/ml) in solvent A (95% acetonitrile + 5% 10mM ammonium acetate). LC-MS/MS analyses were performed using an Agilent 1200 series HPLC-Chip system connected to an Agilent 6490 Triple quadrupole. Hydrophilic interaction chromatography (HILIC) separation was undertaken using a HILIC-chip containing Amide-80 stationary phase (5 µm particle size, 80Å pore size), including a 160 nl trapping column and a 5 µm × 150 mm analytical column (Agilent Technologies Corp., Santa Clara, CA). Separation was achieved using a 19 min gradient of solvent A (95% acetonitrile + 5% 10 mM ammonium acetate) and solvent B (50% acetonitrile + 50% 10 mM ammonium acetate). Gradient was as follows: 100% A for 1.5 min, linear gradient to 90% B over 11.5 min, 90% B for 0.5 min, to 10% B over 0.1 min, 100% A until end of run (total runtime 19 min). Samples were injected (1 μl) through the enrichment column at 4 µl/min. The valve was switched 1.5 min after injection to place the enrichment column in line with the analytical column at a flow rate of 400 nl/min. The mass spectrometer was operated using the following parameters: gas temperature 185°C, gas flow 12 l/min, capillary voltage 1580 V. Multiple reaction monitoring (MRM) transitions (PA to fatty acid fragments) were monitored with a collision energy of 33 V and a fragmentor value of 380 V. Quantification was done by normalizing the area under the curve (AUC) of the chromatographic peak of each MRM transition to the AUC of the internal standard peaks.
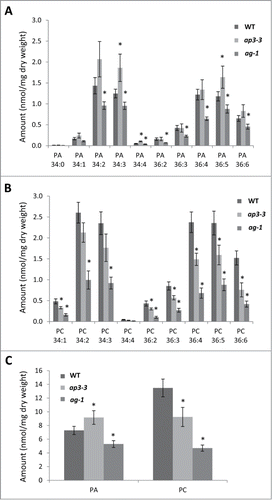
Figure 2. Gene expression analysis for DGKs and PAPs in the flowers of ap3-3, ag-1 and wild type of Arabidopsis thaliana by quantitative reverse transcription-PCR (qRT-PCR) in comparison with published microarray data of wild-type floral organs. (A, B) Transcript levels of DGKs (A) and PAPs (B) in different floral organs of Arabidopsis thaliana by published microarray data (Affymetrix ATH1 Arabidopsis Genome Arrary).Citation12 (C, D) Relative gene expression of 7 DGKs (C) and 11 PAPs (D) analyzed by quantitative qRT-PCR in the flowers of wild type, ap3-3, and ag-1. Data are mean ±SD of 6 replicates including 3 technical replicates of 2 biological replicates. Asterisks indicate significance (p < 0.01) from wild type. Total RNA was extracted from the samples using RNeasy plant mini kit (Qiagen), followed by reverse-transcription to cDNA by SuperScript III reverse-transcription kit (Invitrogen) as described previously.Citation7 qRT-PCR was performed with specific primers designed for each target genesCitation6 using 7500 Real Time PCR System (Life Technologies, Carlsbad, CA). Actin was used as a reference,Citation6 and primer specificity was examined previously.Citation6