Figures & data
Figure 1. Rhizobia perception mediated by plant receptors. Nod Factor (NF) perception is required for infection and nodule organogenesis, but other signaling molecules secreted by bacteria, such as exopolysaccharides (EPS), proteins, cyclic glucans and K antigens and cell wall associated molecules, like the lipopolysaccharide (LPS) have been demonstrated to modulate the infection process. NF and EPS are recognized by LysM receptor-like kinases (LysM RLKs). In particular, NF is perceived by a receptor complex comprising the LysM receptors NFP/LYK3 in M. truncatula or NFR1/NFR5 in L. japonicus. NF signaling activates the root nodule symbiotic pathway and inhibits defense responses. DMI2 and SYMRK are receptors from M. truncatula and L. japonicus that are required for nodulation, but their ligands are unknown. The EPS receptor (EPR3 in L. japonicus, Phvul.002G059500 in common bean) plays a major role in the infection process, suggesting a sequential receptor mediated recognition of NF and EPS. Rhizobial proteins and lectins are recognized by LRR and lectin RLKs, respectively, and have been proposed to regulate defense responses. EPS and LPS negatively regulate two receptors with serine/threonine (Ser/Thr) kinase domains highly similar to the ethylene receptor ETR2 by an unknown process.Citation31 These receptors are regulated by abiotic stresses and hormones. Abbreviations: LRR: leucine rich repeats; NFR: Nod factor receptor; NFP: Nod factor perception; LYK3: LysM receptor kinase 3; DMI2: Does not Make Infection; SYMRK: Symbiosis receptor-like kinase; EPR3: EPS receptor 3; LysM: Lysin motif.
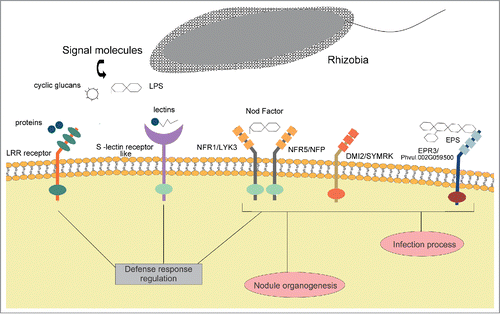
Figure 2. Rhizobium Signal molecules. Representative structures of Nod Factors (NF) exopolysaccarides (EPS) and lipopolysaccharides (LPS). (A) The typical NF backbone consists of 4 or 5 β-1-4 linked N-acetyl-glucosamine residues. NFs are subject to chemical modifications (the position of frequently added groups are indicated with asterisks) by the action of rhizobia nod genes (in bold). The different types of decorations result in a mix of NFs produced by each species of rhizobia. The product of the nodA, nodB and nodC genes participate in the synthesis of the NF backbone. (B) The EPS molecules from S. meliloti Rm1021 are EPS II and EPS I. EPS II is a galactoglucan molecule, whereas EPS I consists of repeating units of octasaccharides modified with acetyl, succinyl and pyruvyl substituents (indicated with asterisks) and is also known as succinoglycan (C) Chemical structure of the LPS from R. etli CE3. LPS is constituted by 3 modules: lipid A, an inner core oligosaccharide and a highly variable O-antigen polysaccharide (OPS). LPSs from rhizobia have variable OPS regions and a number of unique characteristics compared with LPS from enteric bacterial species. The OPS and lipid A regions are key components of the legume-rhizobia interaction.
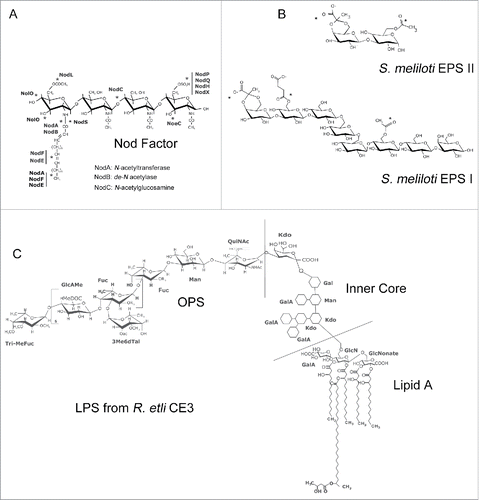
Table 1. Transcriptomic analysis of legumes infected with mutant bacterial strains.
