Figures & data
Table 1. The sequence of primers used in the experiments and the melting temperatures (Tm).
Figure 1. HRF in wt and mutant plants (A) A schematic representation of the luciferase-based recombination substrate and a representative image of recombination events in wt plants. The LUC-based recombination substrate consists of 2 truncated copies of the LUC gene with partial overlapping regions. Recombination events between the regions of overlap (depicted as “U”) result in the restoration of the LUC transgene. Transgene activity can be monitored by spraying plants with luciferin and counting recombination events using a special CCD camera. An example of the CCD camera image is shown. (B) HRF in wt plants and dcl mutants. HRF (an average number of spots per plant with SD) was calculated from 5 independent experiments. The asterisks show the difference in HRF either between wt and mutant plants or between mutant plants (the asterisks over horizontal bars). One asterisks is p < 0.05, two – p < 0.01, three – p < 0.001
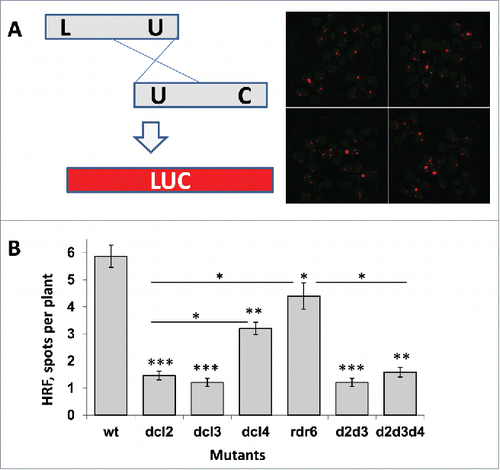
Figure 2. The distribution of recombination events in dcl2 (A), dcl3 (B), dcl4 (C), rdr6 (D), dcl2 dcl3 (E), dcl2 dcl3 dcl4 (F) plants in comparison to wt plants The Y-axis shows the number of plants, whereas the X-axis shows the number of events (spots) per single plant.
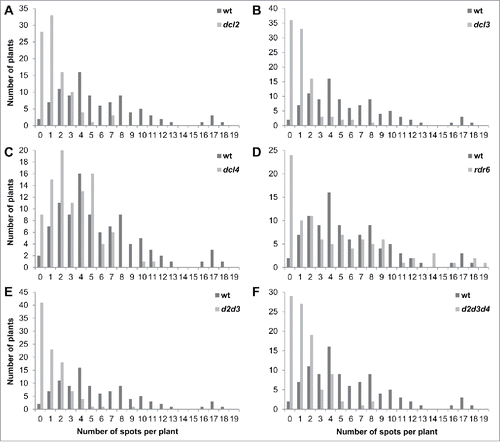
Figure 3. Changes in HRF in response to UVC (A), cold (B), heat (C), flood (D) and drought (E). The Y-axis shows the average (with SDSD) changes in HRF in wt and mutant plants exposed to stress as compared to non-stressed plants. The asterisks show the difference in HRF changes between wt and mutant plants. One asterisks is p < 0.05, two – p < 0.01, three – p < 0.001.
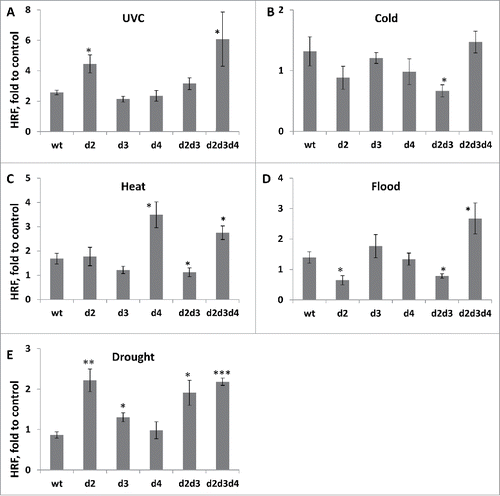
Figure 4. The level of strand breaks in wt and mutant plants SSBs and DSBs levels as measured using the ROPS assay. The data are shown as a percentage to wt (an average with SD), where wt is taken as 100%. The asterisks show a significant difference in the levels of strand breaks between wt and mutant plants. One asterisks is p < 0.05, two – p < 0.01. DSBs levels as measured using the comet assay. The data are shown as an average tail moment (with SD), and all the numbers are related to wt taken as 1. The asterisks (p < 0.01) show a significant difference in DSBs between wt and mutant plants. SSBs and DSBs levels in wt and mutant plants exposed to MMS as measured using the ROPS assay. The data are shown as a percentage to wt control samples (an average with SD), where wt is taken as 100%. The asterisks show a significant difference in the levels of strand breaks either between wt and mutant plants or between MMS treated and control plants (the asterisks over bars). One asterisks is p < 0.05, two – p < 0.01.
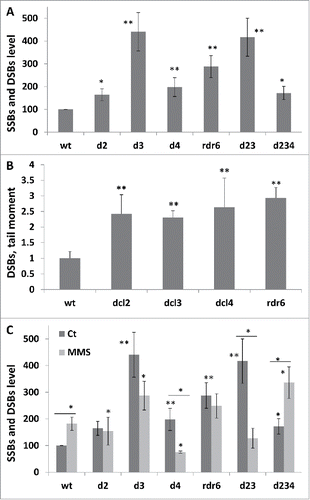
Figure 5. The semi-quantitative RT-PCR (SQ-RTPCR) analysis of the expression of base excision repair, DSB repair and mismatch repaor genes in wt and mutant plants exposed to MMS The steady state RNA levels of AtFPG1 (A), ligase 1 (B), Polλ (C), Rad51 (D), KU70 (E), MSH2 (F) and MSH7 (G) are shown as an average of arbitrary units of intensity calculated from 2 biological and 2 technical repeats (with SD). The asterisks (p < 0.05) show a significant difference between mutants and wt plants.
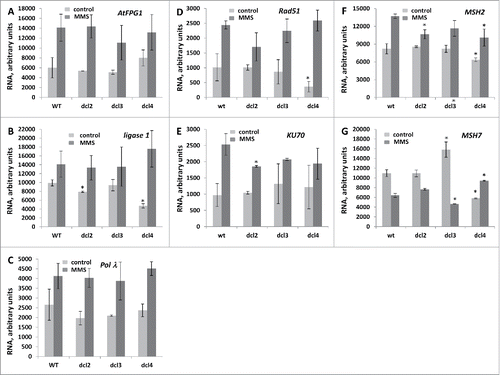
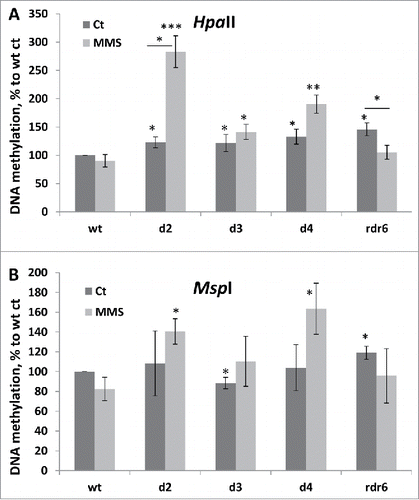
Figure 6. DNA methylation levels in wt and mutant plants in response to MMS. Global genome methylation was analyzed using the cytosine extension assay. Genomic DNA was digested with methylation-sensitive enzymes, HpaII (A) and MspI (B), and the average 3H incorporation (in dpm/μg) was calculated from 4 biological and 2 technical repeats. DNA methylation was calculated using the following formula: methylation = 100/X*100, where X is the radioactive count for a particular sample. DNA methylation is shown in a percentage to wt control plants (wt ct), where wt ct is taken as 100%. The asterisks show a significant difference between mutant and wt samples or between treatment and control samples, where one asterisk is p < 0.05, two – p < 0.01 and three – p < 0.001.