Figures & data
Table 1. Responses of genes encoding ANAC family transcription factors associated with dedifferentiation to AGL15 accumulation in Arabidopsis and soybean. Ranges indicate low and high values from multiple probe sets assigned to soybean loci (Glyma). All results are significant at least at P < 0.05 and for which there is at least a 1.5-fold change.
Table 2. Response of genes encoding WRKY family transcription factors associated with dedifferentiation to AGL15 accumulation in Arabidopsis and soybean. See for remainder of the explanation.
Figure 1. Transcript accumulation patterns for putative soybean orthologs of genes in Arabidopsis data sets corresponding to root and SAM transcriptomes. Data shown are percentage of the total responsive soybean orthologs that are expressed in response to 35Spro:GmAGL15 at 0, 3, and 7 dac (A) or to time in SE induction culture (B) for Jack wt and (C) for 35Spro:GmAGL15). The numbers above the bars indicates the total number of Arabidopsis genes with at least one putative soybean ortholog responsive to 35Spro:GmAGL15 or time in culture.
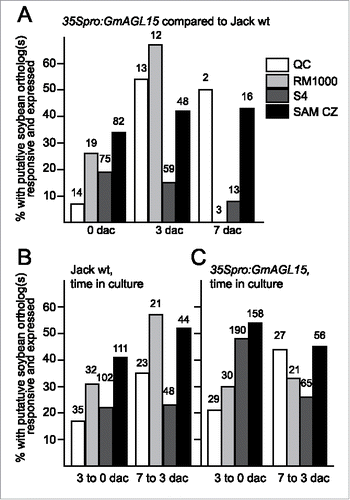
Table 3. Genes from the root QC list (Brady et al.Citation18 Supplemental Table 2, AGL42 tab) with putative orthologs in Glycine max that respond to increased GmAGL15 or to time in culture. Data are presented as the number expressed or repressed out of the total responsive. Some Arabidopsis genes had multiple orthologs in soybean, some of which were expressed and others repressed in the comparisons and these are listed in “both/total.” Percentage of the total genes in each category is in parentheses.
Table 4. Genes responsive to 35Spro:WIND1 (Iwase et al. Citation23) with putative orthologs in Glycine max that respond to increased GmAGL15 or to time in culture. See for the remainder of the explanation.
